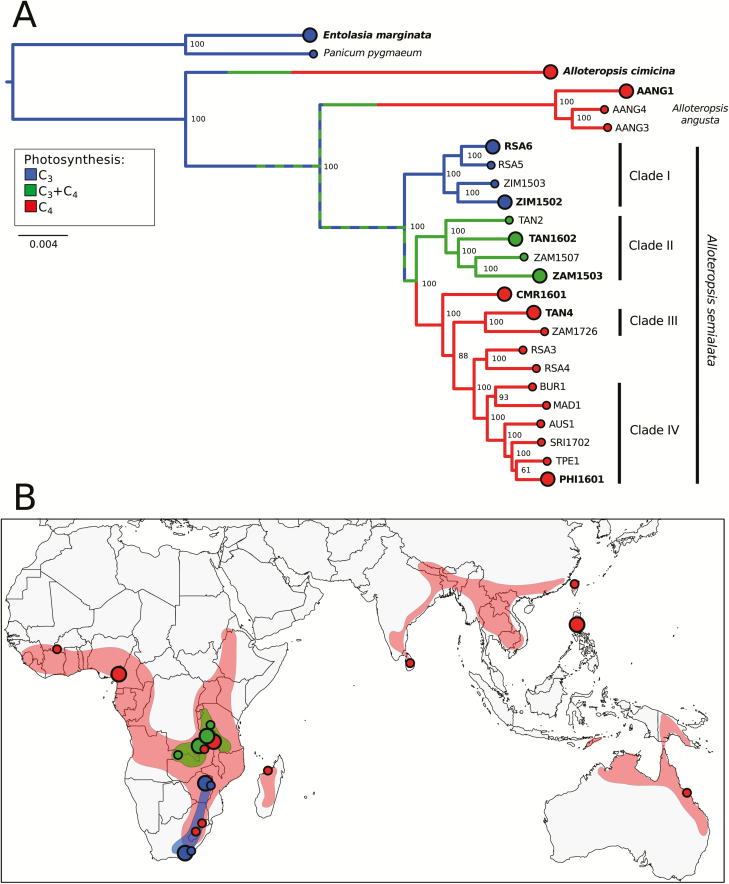
Fig. 1.
Phylogenetic tree inferred from multiple nuclear markers and sampling locations. (A) This phylogeny was inferred under maximum likelihood using transcriptome-wide markers. The scale indicates the number of nucleotide substitutions per site, and bootstrap support values are indicated near nodes. AANG=A. angusta. For A. semialata, population names indicate the country of origin; AUS=Australia, BUR=Burkina Faso, CMR=Cameroon, MAD=Madagascar, PHI=Philippines, RSA=South Africa, TAN=Tanzania, SRI=Sri Lanka, TPE=Chinese Taipei, ZAM=Zambia, ZIM=Zimbabwe. Populations sampled with biological replicates and used for differential expression analysis are indicated by the large circles and bold population names. Nuclear clades from Olofsson et al. (2016) are indicated. Branch colors indicate the ancestral photosynthetic types, based on the transcriptomes and leaf anatomy detailed investigations of Dunning et al. (2017). The hashed green at the base of A. semialata indicates uncertainty between C3 and C3+C4 states. (B) Distribution of A. semialata photosynthetic types and sampling locations, with color codes as in (A). Shadings indicate the approximate ranges of the three photosynthetic types of A. semialata, based on Lundgren et al. (2016).