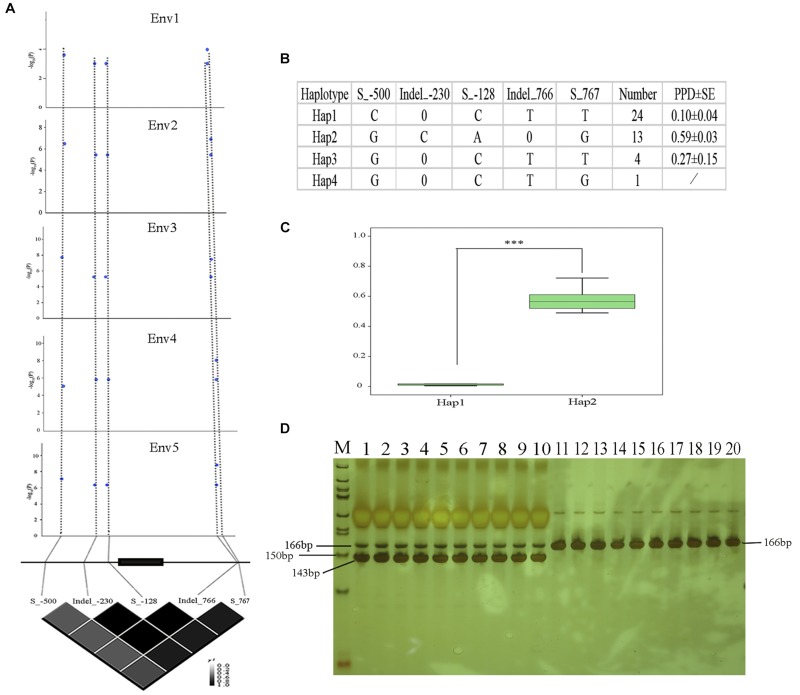
FIGURE 6.
Polymorphisms in Glyma09g06290 are significantly associated with pod dehiscence. (A) Glyma09g06290-based association mapping and pairwise LD analysis. Blue dots represent significant variants (Tassel 5.0, GLM model, P < 0.01). (B) Haplotypes of Glyma09g06290 among 42 soybean accessions; 0 indicates a base deletion. (C) Comparison of PPD between haplotypes Hap1 and Hap2. (D) Products of digestion by gel electrophoresis; M: marker, 50 bp DNA ladder (Tiangen, Beijing, China); 1–10: high-PPD varieties NJAU_C008, NJAU_C014, NJAU_C088, NJAU_C101, NJAU_C121, NJAU_C137, NJAU_C160, NJAU_C180 NJAU_C181, and NJAU_C190. Digested products with size of 166 and 143 bp, 23 bp was not observed because of its small molecular weight; 11–20: low-PPD varieties, NJAU_C054, NJAU_C076, NJAU_C080, NJAU_C082, NJAU_C085, NJAU_C098, NJAU_C165, NJAU_C172, NJAU_C201, and NJAU_C216. Undigested product with size of 166 bp. ∗∗∗Significant at P < 0.001. Two tail t-test was used for statistical analysis. SE, standard error.