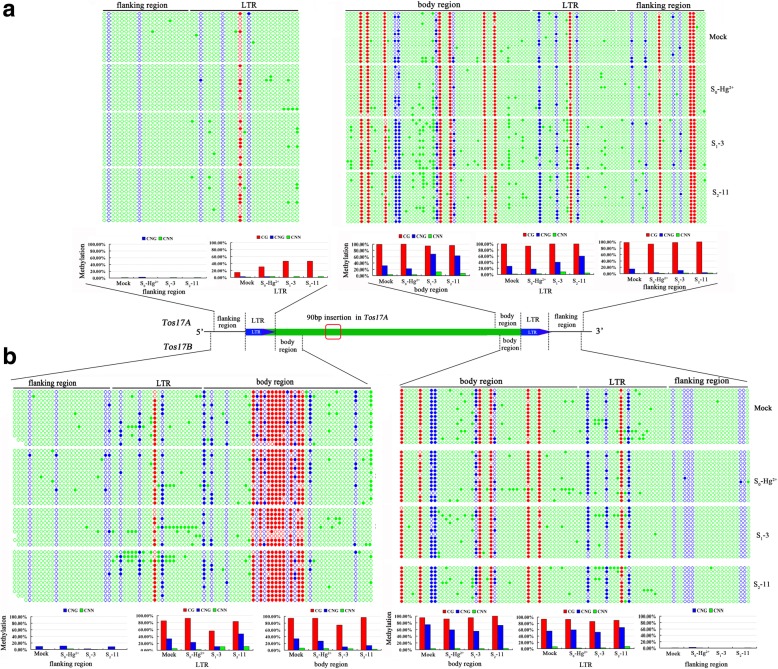
Fig. 4.
DNA methylation status of the Tos17A (a) and Tos17B (b) determined by bisulfite sequencing, respectively, in mock and the Hg-treated S0 plant, and its two successive offspring: S1–3 (S1 generation plant # 3) and S2–11 (S2 generation plant # 11). Specific primers were used on the bisulfite-treated rice genomic DNA to amplify six sites from the two Tos17 (Transposon of Oryza sativa 17) copies in the rice genome (cf. Additional file 2: Table S2). Each copy of Tos17 was amplified from six genomic sites: 3 from the 5′-LTR region (i.e., flanking region, LTR, and body region, expect the body region of Tos17A) and 3 from the 3′-LTR region (i.e., flanking region, LTR, and body region). Subsequently, 10 to 15 clones for each PCR product were sequence analyzed, and the methylation levels per site for each of the three cytosine contexts (CG, CHG, and CHH) were calculated and expressed as a percentage (%). Methylation level was calculated by dividing the number of non-converted (methylated) cytosines with the total number of cytosines underlying a sequenced region. In the picture, each DNA sequences was represented by a string of dots, where filled dots represent methylated cytosines and the open dots represent unmethylated cytosines