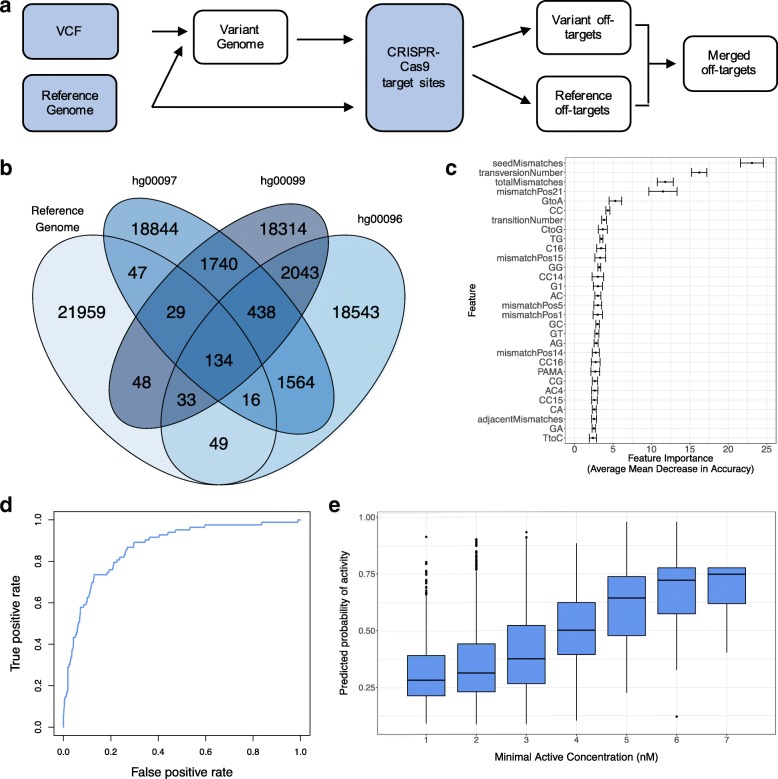
Fig. 1.
Development and testing of the VARSCOT model (a) VARSCOT uses a supplied VCF to produce a variant genome which is searched alongside the reference genome to identify variant off-targets. User supplied files are shown in blue, while files generated by VARSCOT are shown in white. b VARSCOT was used to detect potential off-targets for 100 gRNAs using variant information from three individuals from the 1000 Genomes project. Potential off-targets were compared between individuals and with the hg19 Reference Genome to identify unique targets. c Feature importance for predicting off-target activity. d Receiver Operating Characteristic (ROC) curve of the VARSCOT model tested on the independent Test Dataset filtered for targets with 8 or fewer mis-matches and NGG or NGA PAMs (e) Correlation of the VARSCOT Predicted Probability of Activity with the Minimal Active Concentration of CRISPR-Cas9 used in the Test Dataset