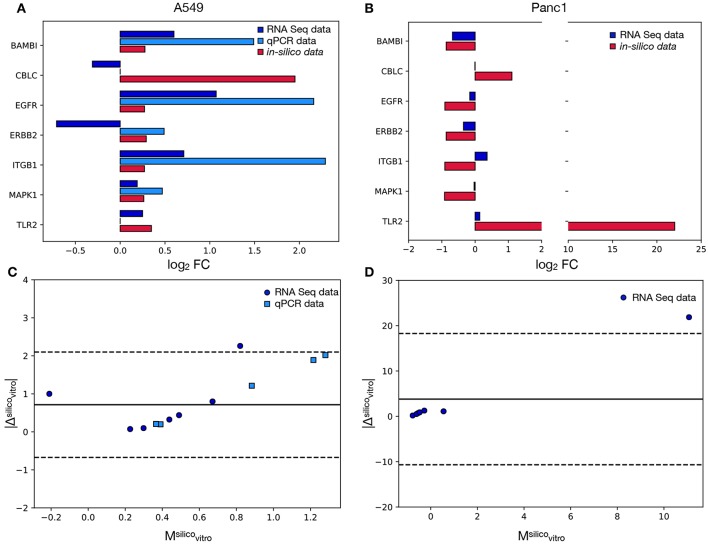
Figure 4.
Comparison between the model's results and the in-vitro data. In (A) the log2FC recorded in A549 cells for each signature marker is reported both for the in-vitro and the in-silico analyses. Different colors are associated with alternative experimental techniques or the model's results. (B) is the same as in (A) but for the Panc1 cells. In this case only the RNA seq data reported in Mishra et al. (2017) were considered. (C) Bland-Altman plot showing the relation between the absolute difference of the in-silico and in-vitro data and their mean. Different colors and symbols identify different comparisons while the solid black line represents the average of the values on the y axis (obtained considering only the RNAseq values). The dotted lines identify the 95% limit of agreement (again computed using only the RNAseq data) and their distance from the solid line is 1.96·σ, where σ is the standard deviation of the absolute difference. (D) Same as in (C) but for the Panc1 cell line.