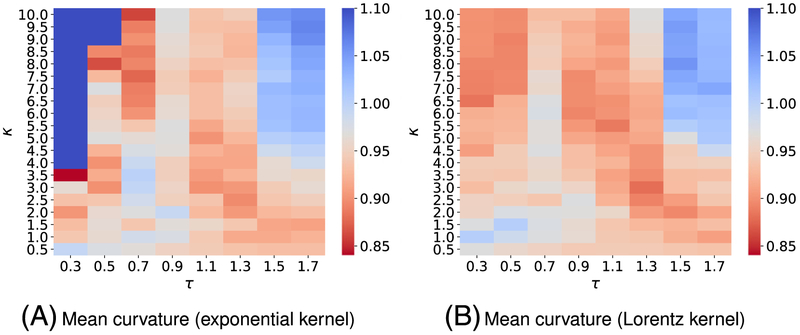
FIGURE B1.
Mean absolute error (MAE) from three-fold cross-validation of on the solvation training set are plotted against different values of τ and κ. The element interactive mean curvatures are utilized for all calculations. The best parameters and median values of MAE for each model are found to be (A) exponential-kernel model: (τ = 0.3, κ = 3.5, MAE = 0.840) and (B) Lorentz-kernel model: (τ = 1.3, κ = 3.0, MAE = 0.880)