Figure 3.
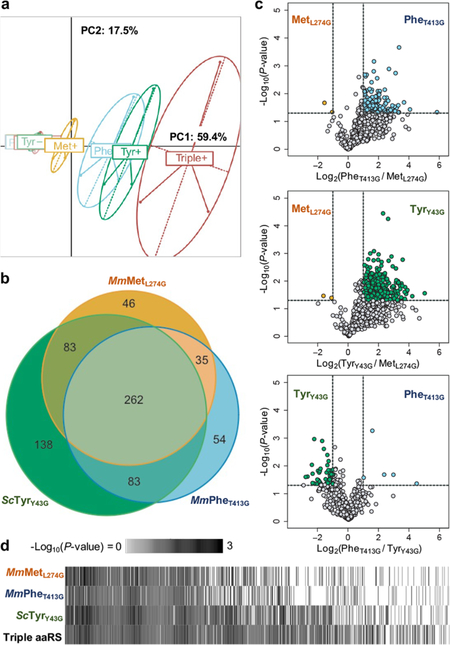
Distinct proteomes labeled by each mutant aaRS. HEK293T cells transfected with equal total amounts of MmMetL274G, ScTyrY43G, and MmPheT413G, or all three aaRSs were incubated with 125 μM of their corresponding azide-bearing ncAA or endogenous amino acids. Lysates were click-enriched on DBCO beads, washed, digested, and TMT labeled before mass spectrometry. n = 3 biological replicates, in technical duplicate, for each condition. (a) Each mutant aaRS differentially labels the HEK293T proteome, as spatially represented by PCA. (b) Each mutant aaRS labels and identifies its own unique set of cell proteins. Most labeled proteins (66%, 463 of 701) are commonly detected across singly expressed mutant aaRSs, increasing confidence in their identification. (c) Among the 463 proteins identified by at least 2 aaRSs, each mutant aaRS exhibits different labeling efficiencies for different proteins. (d) The coexpression of multiple mutant aaRSs (“Triple aaRS”) enhances proteome coverage and detects more proteins with greater confidence, as assessed by P-value. Proteins significantly identified by at least one mutant aaRS were ordered by average -Log10(P-value).