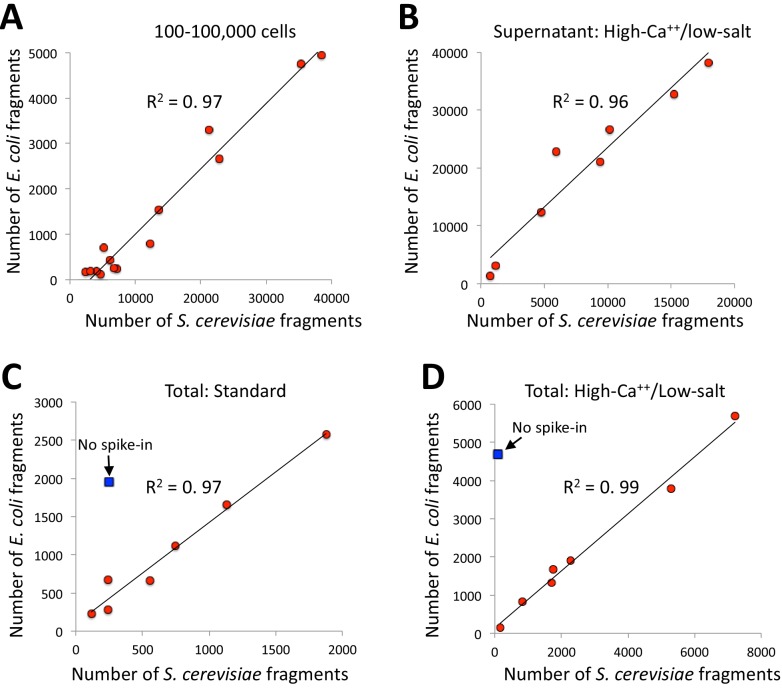
Figure 5. E. coli carry-over DNA of pA/MNase and pAG/MNase can substitute for spike-in calibration.
(A) Fragments from a CUT&RUN K562 cell experiment (GSE104550 20170426) using antibodies against H3K27me3 (100–8,000 cells) and CTCF (1,000–100,000 cells) were mapped to the repeat-masked genome of S. cerevisae and the full genome of E. coli. Standard digestion was followed by supernatant release and extraction. (B) Same as A using antibodies against multiple epitopes of varying abundances, with high-calcium/low-salt digestion followed by supernatant release and extraction. (C) Same as B except using standard digestion conditions and total DNA extraction. The S. cerevisiae spike-in DNA was left out for one sample (blue square). From top to bottom, antibodies are: NPAT Thermo PA5-66839, Myc: CST Rabbit Mab #13987, CTD: PolII CTD Abcam 8WG16, RNAPII-Ser5: Abcam 5408 (mouse), RNAPII-Ser2: CST E1Z3G, CTCF Millipore 07–729, RNAPII-Ser5: CST D9N5I (rabbit), H3K4me2: Upstate 07–030. (D) Same as C except using high-calcium/low-salt digestion and total DNA extraction. From top to bottom, antibodies are: CTCF Millipore 07–729, NPAT Thermo PA5-66839, Myc: CST Rabbit Mab #13987, CTD: PolII CTD Abcam 8WG16, RNAPII-Ser5: Abcam 5408 (mouse), RNAPII-Ser5: CST D9N5I (rabbit), RNAPII-Ser2: CST E1Z3G, H3K4me2: Upstate 07–030.