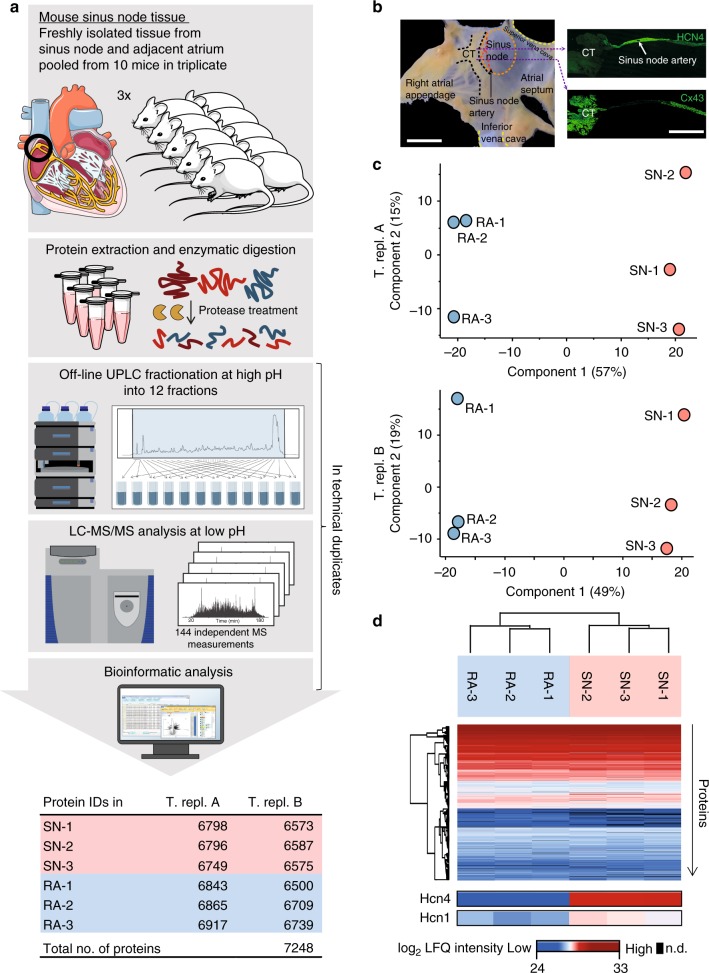
Fig. 1.
High-resolution proteomics measurements of mouse sinus node. a Workflow of proteome measurements. Sinus node (SN) and abutting atrial muscle (RA) was collected from ten mice in triplicates. Tissues were homogenised, followed by protein extraction and digestion. The generated peptides were pre-fractionated by UPLC followed by high-resolution LC-MS/MS analysis and bioinformatic data processing. All steps from UPLC fractionation and onwards were performed in technical replicate experiments, T. repl. A and T. repl. B. The experiments resulted in identification of 7248 proteins. b Left: Mouse right atrial preparation with intact sinus node showing the crista terminalis (CT), sinus node artery, superior and inferior vena cava, atrial septum and the right atrial appendage. Sinus node tissue biopsies were collected from the area demarcated by the dashed orange line and right atrial samples were collected from the atrial appendage. Right: Atrial tissue sections through the sinus node immunolabelled for hyperpolarisation-activated cyclic nucleotide-gated channel 4 (HCN4, top) and connexin 43 (Cx43, bottom) proteins. Note the small size of the sinus node. Scale bars denote 1 mm. c Principal component analysis (PCA) of proteomics data for technical replicates A and B shows that 49–57% of the variation in the dataset is explained by differences in protein expression between sinus node (red) and atrial muscle (blue). d Unsupervised hierarchical clustering of protein intensities shows grouping of sinus node and atrial muscle into distinct clusters. Data for HCN proteins is highlighted at bottom. On the colour scale, blue and red indicate low and high protein expression level, respectively. UPLC: ultra-high pressure liquid chromatography; MS: mass spectrometry; LC: liquid chromatography. Source data is provided in Supplementary Data 1