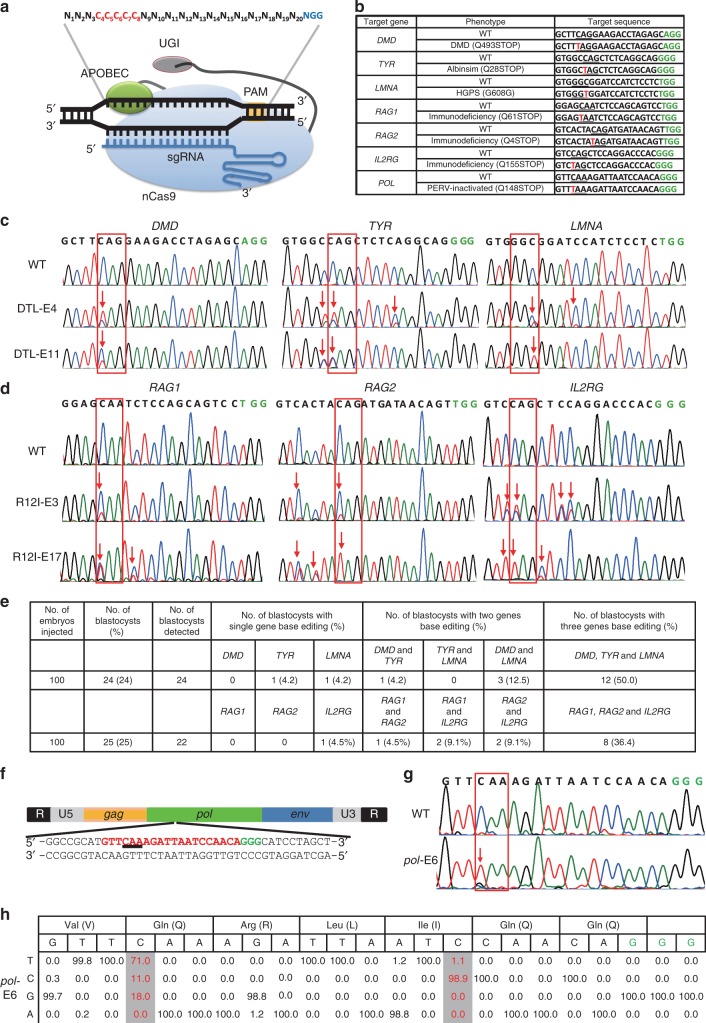
Fig. 1.
The BE3 system can simultaneously induce C-to-T base editing at multiple genes/loci of porcine embryos. a Schematic of BE3-mediated C-to-T base editing. b Target-site sequences of DMD-, TYR-, LMNA-, RAG1-, RAG2-, IL2RG-, and pol-sgRNA. Target sequence (black), protospacer adjacent motif (PAM) region (green), target sites (red), and mutant amino acid (underlined). WT wild-type. c Sanger sequencing results of embryo-4# and 11# injected with DMD-sgRNA, TYR-sgRNA, and LMNA-sgRNA. The red box shows the successful C-to-T substitutions at target sites. d Sanger sequencing results of embryo-3# and 17# injected with RAG1-sgRNA, RAG2-sgRNA, and IL2RG-sgRNA. e Summary of multiple sites base editing by BE3 in porcine embryos. f Schematic of the PERV gene structure. One sgRNA targeting the catalytic region of the PERV pol gene was designed. The codon to be modified is underlined. The targeting sequence is in red and the PAM region is in green. g Representative sequence chromatogram of the target site of the pol gene from injected embryos 6#. Red box shows the successful C-to-T substitutions at target sites. h Nucleotide substitution frequencies mediated by BE3 and pol-sgRNA were measured in the injected embryos 6# by targeted deep sequencing