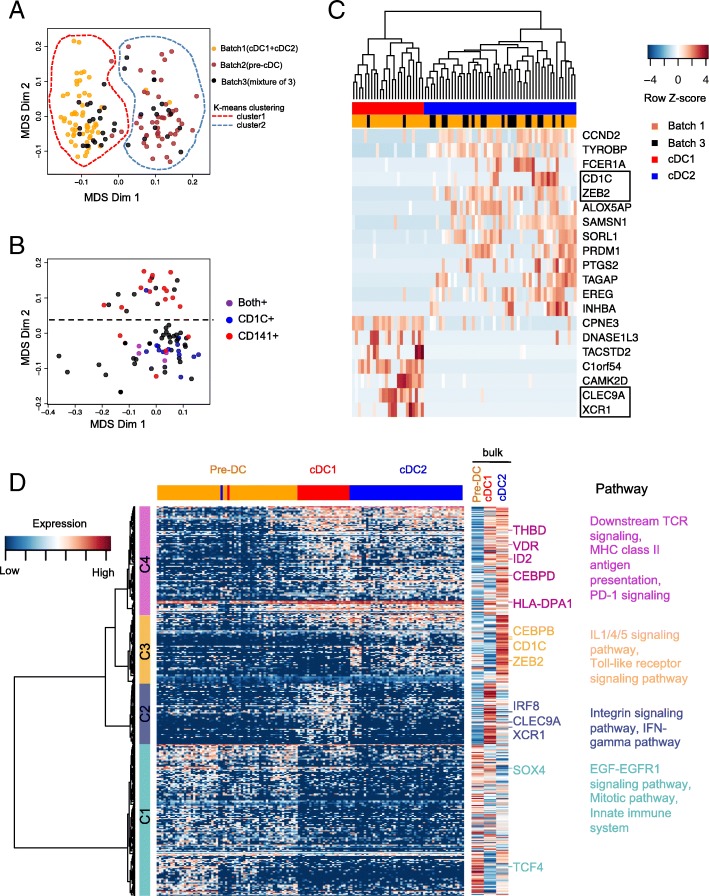
Fig. 2.
Cell type identification of the single cells from mixture with MDS based on global transcriptome. a Multidimensional scaling (MDS) plot for the cells that passed quality control, batch1 (yellow, cDCs), batch2 (brown, pre-cDCs) and batch3 (black, mixture of cDCs and pre-cDCs), where the input distance was adjusted for dropout rate with SCDE. The cells can be separated into 2 clusters by k-means clustering: cluster1 for cDCs and cluster 2 for pre-cDCs. b The clustering pattern of cluster1 identified in panel a with MDS. Each cell is colored with binary expression of CD141(red), CD1c (blue), both (purple) or none (black). c Heatmap of the top 20 differentially expressed genes derived by SCDE by comparing the upper and lower groups of cDCs in panel b. The top dendrogram was generated with hierarchical clustering. The two column color bars represent batch and cell type information, respectively. Genes surround by black box are known key genes for cDC differentiation. d Transcriptional signature of pre-cDC, cDC1 and cDC2 population. Three hundred eighty genes shown in rows were differentially expressed in at least two populations of pre-cDC, cDC1 and cDC2 computed with ANOVA (false-discovery rate (FDR) < 0.05) and can be classified into 4 clusters(C1–4) indicated by the row slide color. Bulk expression of the three cell types for these genes after averaging over 3 biological replicates is also shown on the right. Right margin also shows the predicted upstream regulators and enriched pathways for each gene cluster. Single cells in the columns were ordered by the hierarchical clustering result based on the 380 genes