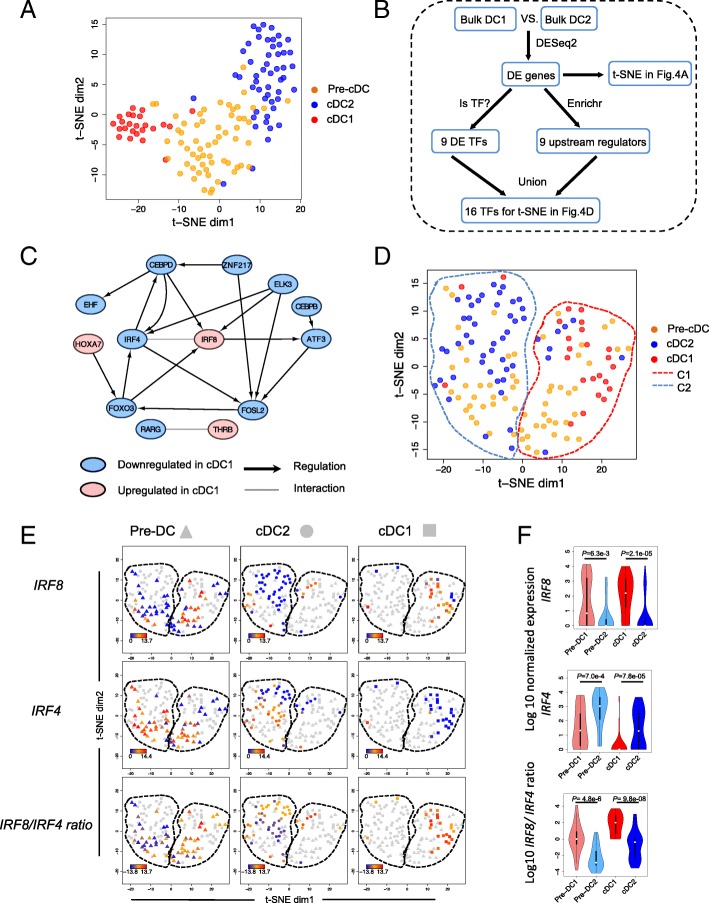
Fig. 4.
Pre-cDC subpopulation pattern in human is implicated by differentially expressed TFs between cDC1 and cDC2. a t-SNE plot of all the single cells with the full set of differentially expressed genes between bulk cDC1 and cDC2. b Flowchart of how to identify the MR TFs. c Regulatory network of the 13 out of 16 MR TFs based on the database of transcription factor and target relationship in human hematopoietic lineages from Neph et al. [25] complemented by String database [26]. d t-SNE plot of all the single cells with MR TFs that are differentially expressed between bulk cDC1 and cDC2. The cells were clustered into two groups C1 and C2 by k-means clustering. e Expression level of IRF8, IRF4 and IRF8/IRF4 expression ratio for each single cell. The three rows represent IRF8, IRF4 and IRF8/IRF4, respectively. The columns represent pre-cDC, cDC2 and cDC1, respectively. For example, the IRF8 expression level from low to high in pre-cDCs is represented from blue, orange to red, and the other cDC single cells are colored in gray. f Violin plot of IRF8 expression, IRF4 expression and IRF8/IRF4 expression ratio in single cell populations: pre-cDC1, pre-cDC2, cDC1 and cDC2. P values indicated between pre-DC1 and pre-DC2 and between cDC1 and cDC2 are from Wilcoxon Rank Sum test