Abstract
Background
Sucrose is an attractive industrial carbon source due to its abundance and the fact that it can be cheaply generated from sources such as sugarcane. However, only a few characterized Escherichia coli strains are able to metabolize sucrose, and those that can are typically slow growing or pathogenic strains.
Methods
To generate a platform strain capable of efficiently utilizing sucrose with a high growth rate, adaptive laboratory evolution (ALE) was utilized to evolve engineered E. coli K-12 MG1655 strains containing the sucrose utilizing csc genes (cscB, cscK, cscA) alongside the native sucrose consuming E. coli W.
Results
Evolved K-12 clones displayed an increase in growth and sucrose uptake rates of 1.72- and 1.40-fold on sugarcane juice as compared to the original engineered strains, respectively, while E. coli W clones showed a 1.4-fold increase in sucrose uptake rate without a significant increase in growth rate. Whole genome sequencing of evolved clones and populations revealed that two genetic regions were frequently mutated in the K-12 strains; the global transcription regulatory genes rpoB and rpoC, and the metabolic region related to a pyrimidine biosynthetic deficiency in K-12 attributed to pyrE expression. These two mutated regions have been characterized to confer a similar benefit when glucose is the main carbon source, and reverse engineering revealed the same causal advantages on M9 sucrose. Additionally, the most prevalent mutation found in the evolved E. coli W lineages was the inactivation of the cscR gene, the transcriptional repression of sucrose uptake genes.
Conclusion
The generated K-12 and W platform strains, and the specific sets of mutations that enable their phenotypes, are available as valuable tools for sucrose-based industrial bioproduction in the facile E. coli chassis.
Electronic supplementary material
The online version of this article (10.1186/s12934-019-1165-2) contains supplementary material, which is available to authorized users.
Keywords: Escherichia coli, Renewable feedstocks, Sucrose, Adaptive laboratory evolution, Platform strains
Introduction
There is a significant interest in the utilization of renewable carbon feedstocks for bioprocesses due to both environmental and economic factors. Sucrose from sugarcane can serve as a renewable carbon source as it originates from a cheap abundant feedstock source which can lower carbon source costs [1, 2], and it can be readily utilized by biological systems in fermentation processes either from the raw source as sugarcane juice [3] or from the refined byproduct of the sugar industry as molasses [4, 5].
The ability of Escherichia coli, an industrial biotechnology workhorse [6–8], to utilize sucrose as a sole carbon source depends on the specific E. coli strain used [9]. E. coli W is the only well-known E. coli strain generally regarded as safe that can utilize sucrose as a carbon source and can grow robustly on it when compared to other carbon sources, such as glucose [10]. The genetic basis and molecular control of sucrose metabolism in E. coli W (as well as E. coli EC3132) have been characterized [11, 12] and this knowledge is the basis for constructing sucrose utilizing E. coli strains for the current study. Engineering non-native sucrose utilization into additional E. coli strains has been investigated, but in all cases, the generated strains were characterized by slow growth rates on sucrose as compared to glucose. Such low growth rates would limit productivity in industrial processes, a key component for a viable bioprocess [13, 14]. Most of the previous approaches to engineer E. coli to use sucrose as a carbon source were hampered by slow growth rates and phenotypic instability due to unstable plasmid systems that may require antibiotic addition for stability [15] or due to a high burden exerted on the cell by high copy number plasmids [11, 16–18]. For example, the K-12 strains generated by Tsunekawa et al. using chromosomal integration grew with very slow rates on sucrose and random mutagenesis was required to improve the growth rate modestly [19]. Another approach examined was to chromosomally integrate sucrose utilization genes from E. coli W into an E. coli K-12 strain [20]. The K-12 strain generated was able to grow on sucrose, but the growth rate was 30% lower than that on glucose. Thus, there is still a need to efficiently engineer sucrose metabolism in multiple E. coli strains, each of which have their own strain-specific advantages for host selection for viable bioprocesses [6]. Such host selection factors to consider include product tolerance, phage resistance, the native metabolic flux distribution either towards a native or heterologous production pathway, transformation efficiency, convenience to perform metabolic changes, and ease to scale-up [21].
In order to demonstrate the ability to generate additional E. coli strains that can efficiently consume sucrose, genetic constructs were designed based on the chromosomally encoded sucrose catabolism operon, csc, from E. coli W and then subjected to adaptive laboratory evolution (ALE). The csc gene cluster containing the cscB gene that encodes sucrose permease, cscK that encodes a fructokinases, and cscA that encodes sucrose-6-phosphate hydrolase (invertase), was integrated into the E. coli K-12 MG1655 chromosome [11] (Fig. 1). The cscR gene that encodes for a csc-specific repressor and negatively controls the expression of the csc regulon was not integrated to allow constitutive expression. The engineered strain was then optimized using ALE to generate multiple evolved E. coli strains able to metabolize sucrose with fast growth comparable to that on glucose. Additionally, the approach to analyze multiple independent populations and multiple independent isolates from each population made it possible to effectively reveal key causal mutations by comparing the independent lineages and focusing on instances of parallel evolution. This approach also likely identified a broader landscape of mutations (e.g., multiple alleles of the same gene) as compared to sequencing a single lineage and reverse engineering all of the mutations to find causality. The ALE approach used in the current work used an automated platform with several parallel replicates as previously described [22, 23]. Overall, cells were repeatedly grown in a batch aerobic cultivation mode with passage in the exponential phase, which applies selection pressure for rapid growth rate. At the end of the evolution, populations and clones from the ALE derived endpoints were characterized in terms of their genome sequence, growth rate, carbon uptake rate, and growth yield. Furthermore, the evolved strains were characterized on sugarcane juice as a substrate demonstrating their ability to use this renewable feedstock efficiently.
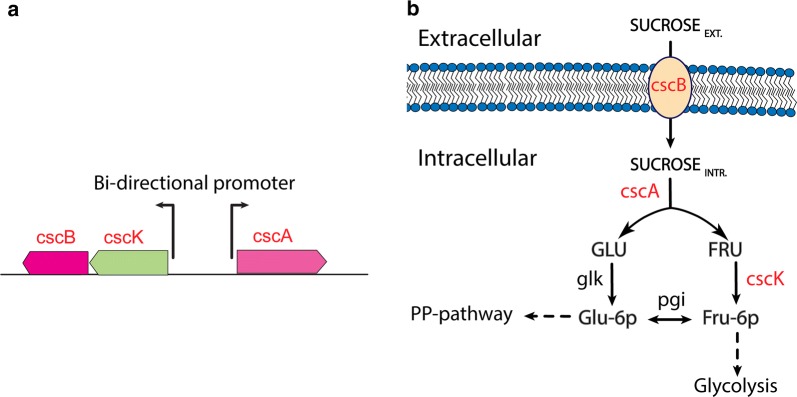
Fig. 1.
Schematic presentation of sucrose utilization mechanism with gene cassette integrated into E. coli K-12 MG1655 and route of sucrose entering into the central carbon metabolism. a The sucrose integrated cassette csc consists of cscA, cscK, and cscB. CscBK and cscA expression is derived by a bi-directional promoter. b Sucrose utilization starts with uptake of sucrose across the cell membrane by cscB (sucrose permease). Inside the cell sucrose is split into glucose and fructose by cscA (invertase). Glucose and fructose are then phosphorylated by glucokinase (glk) and fructokinase (cscK) respectively. Glucose-6-phosphate can be converted to fructose-6-phosophate by the isomerase (pgi) or enter directly into the pentose phosphate pathway (PP-pathway) or Entner-Doudoroff pathway (ED-pathway); fructose-6-phosphate enters the glycolysis directly
Methods and materials
Media and reagents
M9 minimal medium
M9 sucrose medium contained 20 g/L sucrose unless stated otherwise, 1× M9 salts, 2 mM MgSO4, 100 μM CaCl2 and 1× trace elements and Wolfe’s vitamin solution. M9 salts, trace elements and vitamins were prepared in concentrated stocks. Stock M9 salts solution consisted of 10 × 68 g/L Na2HPO4 anhydrous, 30 g/L KH2PO4, 5 g/L NaCl, and 10 g/L NH4Cl dissolved in Milli-Q filtered water. M9 trace elements stock was a 2000x solution with composition of 3.0 g/L FeSO4·7H2O, 4.5 g/L ZnSO4·7H2O, 0.3 g/L CoCl2·6H2O, 0.4 g/L Na2MoO4·2H2O, 4.5 g/L CaCl2·H2O, 0.2 g/L CuSO4·2H2O, 1.0 g/L H3BO3, 15 g/L disodium ethylene-diamine-tetra-acetate, 0.1 g/L KI, 0.7 g/L MnCl2·4H2O and concentrated HCL dissolved in Milli-Q filtered water. Vitamin mix and trace elements concentration was [1×] in the M9 medium.
Sugarcane juice medium
Raw sugarcane juice was collected from a sugar cane mill (Enterprise Factory, Patoutville, Louisiana, USA) and transported to the University of Illinois Energy Farm where it was stored at − 80 °C until required for further use. Sugarcane juice (SCJ) for the cultivation medium was prepared by centrifugation in order to remove solid particles (rpm ≥ 10,000×g for 20 min at 4 °C) then filter sterilized with 0.2 µm pore size filter. Later, Sugarcane juice (SCJ) medium was prepared by mixing clarified sugarcane juice with M9 salts (final concentration 1×), 2 mM MgSO4, 100 μM CaCl2 and 1× trace elements and Wolfe’s vitamin solution. HPLC analysis of the clarified sugar cane juice showed the following sugar composition (g/L): sucrose 231.8, glucose 3.5; fructose 3.0.
Strains constructions with cscBKA cassette using homologous recombination
General recombinant DNA techniques were applied according to standard protocols for one-step cloning and chromosomal integration of DNA (pOSIP), Additional file 2: Text file S1 [24, 25]. PCR products were purified using plasmid miniprep kit, QIAprep Spin Miniprep Kit. Here, the integration site of csc DNA sequence was into E. coli 186 attB site using pOSIP-KO integration module. The integration site of DNA sequence into bacterial chromosomes was selected based on the available integration sites. Csc gene cluster (cscK, cscB, cscA) (Fig. 1a) was amplified from donor strain, E. coli W using primers; Forward primer 5′-ATGCATCUGGGATATAGAGCTATCGACAACAACCG-3′ and Reverse primer 5′-AGAGGGCUTTATGTTAACCCAGTAGCCAGAGTGCTC-3′. E. coli K-12 construct with csc cassette integrated was abbreviated to MGcscBKA. MGcscBKA derivative with a variant SNP in cscB Q353H was abbreviated MGcscBKAp [16].
Adaptive laboratory evolution of developing improved fitness in start strains
Three biological replicates from each of E. coli strains; two genetic constructs of K-12 MG1655 with csc gene cassette MGcscBKA and MGcscBKAp and E. coli W strain, were grown overnight in M9 minimal media with 20 g/L sucrose. On the next day 150 µL of the overnight culture was passed into a new fresh 30 mL tube filled with a total working volume of 15 mL M9 medium (i.e., a 1:100 ratio) with 20 g/L sucrose as the sole carbon source. Bacterial cells were serially passaged during exponential growth phase for approximately 40 days using an automated liquid-handler platform as described by Lacroix et al. [22]. The liquid-handler platform was utilized to automate executing multiple evolution experiments at the same time. It has the capacity to aspirate cell cultures when passing from a mature culture tube to a freshly filled tube based on a pre-defined media recipe in a sterile strictly-controlled platform, in an effort to keep the cells growing and passing at the exponential phase of growth. The commonly encountered exponential phase was from the time of the inoculation to an approximate optical density of 600 nm (OD600nm) of 1.3–2.0 in order to keep the cells under a constant selection pressure. Cells were cultured in a heat block at 37 °C with magnetic stir bar for full aeration at 1200 rpm. Periodically, OD600nm was measured at a time determined by a predictive script and once OD600nm reached approximately 1.3 using Sunrise plate reader (Tecan, Männedorf, Switzerland), 150 µL was passed into a tube with a 15 mL working volume of fresh media. The common conversion factor between the plate reader used and the Benchtop spectrophotometer is 4.2. This process was repeated until a significant increase in fitness was achieved. Periodically, glycerol cryogenic stocks were prepared and stored at − 80 °C for any culture restarting. Endpoint population samples were streaked on agar plates to select single clones for whole genome resequencing.
Genome sequencing and mutation calling of the ALE derived strains
Overall, there were six endpoint populations for each of K-12 MG1655 constructs and 3 endpoints populations from E. coli W were selected along with clones derived from for whole genome re-sequenced in order to reveal their underlying genotypes. Genomic DNA was extracted from overnight cultures at the stationary phase of growth using PureLink® Genomic DNA extraction kits (Invitrogen, CA). The quality of extracted DNA was assessed by using a Nanodrop spectrophotometer. Concentration of the extracted DNA was quantified using Qubit ds-DNA high sensitivity assay. Paired-end re-sequencing libraries were generated using a 300 cycle (150 bp × 2) kit from Illumina (San Diego, CA) with loading concentration of 1.2 pico-Molar on Illumina Nextseq sequencer (Model 550). Mutation finding was performed using a pipeline as described in Phaneuf et al. [26] based on Breseq version 0.30.1 [27] to map sequenced reads to the reference strain (NCBI accession number NC_000913.3, K-12 MG1655 and NC_017664, W). The average coverage for each of the resequenced samples was over 25x. For population samples sequenced, mutations were reported if they were over 20% frequency unless they were found in a clone isolated from a given population sample. In this case, the frequency of a clone mutation was reported, if found, in the population (see Additional file 1: Data file S1).
Validation of mutations causality in rpoB, rpoC and pyrE-rph genes
Identified causal mutations found in the current study (see mutation analysis results) and the control reference ALE experiment by LaCroix et al. [22] on glucose (GLUALE) were used to check causality in the identified genes or genetic regions on either sucrose and glucose minimal media. Briefly, the identified key single point mutations (SNPs) in open reading frames (ORF) mainly for rpoB, rpoC genes and the unique deletion in the intergenic region between pyrE/rph were used to check the causality of mutations in these specified genetic regions with sucrose for growth increase from the two experiments. Accordingly, selected isolates from ALE derived clones on sucrose which harbor any single mutation or double mutations in these genes were selected as well. Fitnesses increase comparison relative to the starting strain were examined to investigate the effect/essentiality of the key causal mutations for fast growth on either sucrose or glucose as a sole carbon source. The same recombinant DNA technique used to generate MGcscBKA constructs with the csc regulon was applied here to generate GLUALE constructs with a sucrose utilization cassette (GLUALE_csc constructs).
Extracellular metabolites and physiological properties
Cultures of the re-sequenced clones were inoculated from stationary phase overnight cultures into media M9 containing sucrose under the same conditions as the ALE experiment. Samples were aliquoted over the growth curve to measure optical density OD600nm and collect extracellular metabolites. Extracellular metabolites were collected as supernatant from each growing culture using 0.2 µm filter to remove the cells. Supernatants were collected and saved at − 20 °C for subsequent chromatographic analyses. Concentration of sugars (glucose, fructose and sucrose) beside other organic acids were analyzed using high performance liquid chromatography (HPLC) column (UltiMate 3000, Thermo-Fischer Scientific, Waltham, Massachusetts, USA). The metabolites were separated using an Aminex HPX-87H ion exclusion column (Bio-Rad, Hercules, California, USA) and were isocratically eluted at 30 °C, with a flow rate of 0.6 mL/min, using a 5 mM sulfuric acid solution as mobile phase. The refractive index (RI) detector was selected for detection. Sample concentrations were quantified by comparing to a standard curve of known concentrations. Substrate uptake and metabolites excretion rates were calculated from multiplying the growth rate and the slope of a linear regression of gram dry cell weigh (gCDW) versus the substrate or products concentration. The biomass yield at the steady state (YX/S_ss) was calculated as the quotient of the growth rate and the sucrose uptake rates during the exponential growth phase.
Results
Evolution of multiple E. coli strains to grow rapidly on sucrose minimal media
ALE was utilized to generate strains with improved fitness utilizing sucrose as a sole carbon source. Three different E. coli starting strains were used; two engineered K-12 MG1655 strains with sucrose utilization enabling csc constructs and wild-type E. coli W. The two constructs inserted into the K-12 MG1655 host genome differed by a SNP mutation in the cscB gene, resulting in the residue change Q353H, and were labeled as MGcscBKA and MGcscBKAp (see Additional file 2: Text S1 for a detailed description). The Q353H derivative of cscB has been demonstrated to display an increased sucrose uptake rate in E. coli EC3132 [16], thus it was reasoned that it may have an impact when heterologously expressed in K-12 MG1655.
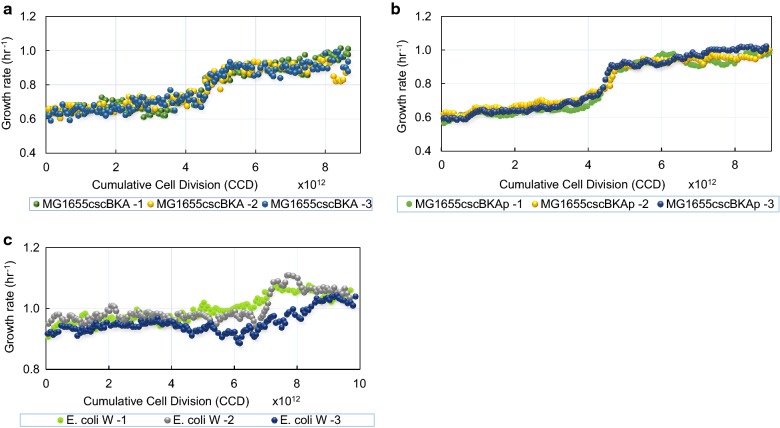
Three independent populations of each starting strain were evolved in batch and in parallel under the strict selection pressure of continuous exponential growth for approximately 40 days (Table 1, Fig. 2a–c). Termination of the ALE experiment was determined based on two key parameters [28]; the delta change in growth rate (Δμ) and the passage size. Therefore, the experiments were terminated when the parallel replicates showed a small to no delta change in the growth rate similar to the control ALE on glucose [22], with the passage volume equal to 1%. Each of the replicate evolutions underwent between approximately 8.65 × 1012 to 9.87 × 1012 cumulative cell divisions (CCD, Table 1). The use of CCD has been demonstrated as a useful timescale for ALE experiments as a time coordinate as it accounts for variability due to a varying number of cells passed serially from one flask to the next [29].
Table 1.
Properties of the ALE experiments end point populations
| ALE experiment | Strain (population) | Total CCD x1012 | Ratio of fitness increase to start strain |
|---|---|---|---|
| K-12 MGcscBKA | 1 | 8.65 | 1.72 |
| 2 | 8.75 | 1.46 | |
| 3 | 8.94 | 1.61 | |
| K-12 MGcscBKAp | 1 | 9.02 | 1.48 |
| 2 | 9.02 | 1.47 | |
| 3 | 8.85 | 1.48 | |
| E. coli W | 1 | 9.71 | 1.29 |
| 2 | 9.78 | 1.31 | |
| 3 | 9.87 | 1.22 |
Ratios of fitness increase were determined from growth rates that were calculated based on the last three flasks during exponential growth. CCD, cumulative cell divisions
Fig. 2.
Plots of population fitness (i.e., growth rate) trajectories during the ALE experiments on sucrose. Three biological replicates of each of the E. coli strains: a K-12 MG1655cscBKA, b K-12 MG1655cscBKAp, and c W were evolved on 20 g/L sucrose minimal medium. Shown are the growth rates versus cumulative cell divisions (CCD) for the three biological replicates for each strain
The observed growth rate trajectories during the ALE experiments for each of the evolved populations are shown in Fig. 2 for the MGcscBAK, MGcscBKAp, and E. coli W strains. Each population displayed an increase in growth rate over the starting strain (Table 1). The growth rate increases were 1.49 ± 0.098-, 1.75 ± < 0.01-, 1.25 ± 0.036-fold faster than the starting strains for the MGcscBKA, MGcscBKAp and E. coli W strains, respectively (standard deviation, n = 3). There was one observed fitness jump (i.e., increase in growth rate) for all the populations across all independent ALE replicates. The two similar K-12 MGcscBKA and MGcscBKAp strains evolved to relatively similar growth rates, with similar dynamics along the course of the ALE, whereas the fold increase in fitness for E. coli W was small given its initial faster wild-type growth rate.
Physiological characterization of evolved clones
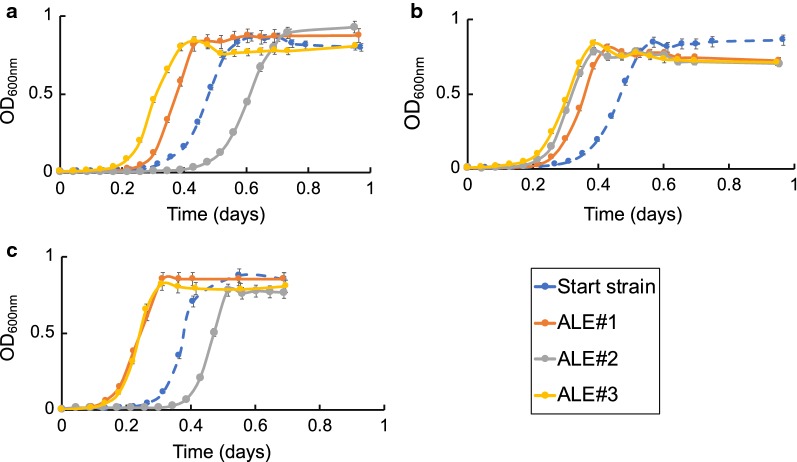
Clones from the endpoint populations of each of the nine replicate ALEs were isolated and characterized in terms of growth rate and compared to the starting strains to understand the enhanced evolved phenotype. One clone isolated from each replicate endpoint population was analyzed, three clones for each of the three replicates of MGcscBAK, MGcscBKA, and E. coli W. The increases in growth rates on sucrose M9 were between 1.17 and 1.57-fold for MGcscBKA clones, 1.25–1.38-fold for MGcscBKAp, and 1.06–1.22-fold for E. coli W, Table 2. Surprisingly, there was one isolated clone that had the same fitness, i.e. growth rate, as the starting strain from MGcscBKA (the ‘#2’ clone) (Fig. 3). However, given that there were two additional clones from this starting strain from the additional two independent replicates with a significant improvement in fitness, additional clones were not examined.
Table 2.
Physiological data for the evolved isolates on sucrose M9 medium and sugarcane juice M9 minimal medium (SCJ medium)
| Clone/construct | Clone ID | M9 Sucrose medium | SCJ medium | |||||
|---|---|---|---|---|---|---|---|---|
| Growth rate, μ (h−1) on M9 2% sucrose | Growth rate, μ (h−1) | Final density (gCDW/L) | Sucrose uptake rate (mmol gCDW−1 h−1) | Acetate production rate (mmol gDW−1 h−1) | Biomass yield, YX/S, (gCDW g of sucrose−1) | Uptake rate fold increase vs start strain | ||
| K-12 MGcscBKA | Starting strain | 0.54 ± 0.01 | 0.68 ± 0.03 | 1.47 ± 0.04 | 7.56 ± 0.96 | 4.20 ± 0.20 | 0.28 ± 0.01 | 1.00 |
| 1 | 0.85 ± 0.01 | 0.88 ± 0.02 | 1.91 ± 0.03 | 8.71 ± 0.2 | 4.73 ± 0.11 | 0.30 ± 0.01 | 1.15 | |
| 2 | 0.63 ± 0.01 | 0.68 ± 0.02 | 1.52 ± 0.04 | 6.54 ± 0.23 | 5.67 ± 0.54 | 0.30 ± 0.01 | 0.86 | |
| 3 | 0.83 ± 0.01 | 0.86 ± 0.02 | 1.68 ± .0.03 | 7.84 ± 0.18 | 5.42 ± 0.13 | 0.32 ± 0.01 | 1.04 | |
| K-12 MGcscBKAp | Starting strain | 0.64 ± 0.01 | 0.73 ± 0.01 | 1.48 ± 0.02 | 7.01 ± 1.9 | 4.99 ± 0.25 | 0.30 ± 0.01 | |
| 1 | 0.86 ± 0.01 | 0.91 ± 0.01 | 1.38 ± 0.02 | 9.89 ± 0.19 | 6.83 ± 0.01 | 0.27 ± 0.01 | 1.41 | |
| 2 | 0.88 ± 0.01 | 0.88 ± 0.01 | 1.44 ± 0.02 | 9.50 ± 0.01 | 8.27 ± 0.01 | 0.27 ± 0.01 | 1.36 | |
| 3 | 0.80 ± 0.01 | 0.82 ± 0.01 | 1.51 ± 0.01 | 8.98 ± 0.11 | 7.30 ± 0.09 | 0.27 ± 0.01 | 1.28 | |
| E. coli W | Starting strain | 0.90 ± 0.02 | 0.97 ± 001 | 1.81 ± 0.05 | 9.88 ± 0.35 | 6.60 ± 0.49 | 0.29 ± .01 | |
| 1 | 1.10 ± 0.01 | 1.06 ± 0.05 | 1.68 ± 0.06 | 10.5 ± 0.47 | 15.90 ± 0.68 | 0.29 ± 0.012 | 1.06 | |
| 2 | 0.95 ± 0.01 | 0.96 ± 0.02 | 1.74 ± 0.05 | 14.10 ± 1.0 | 6.50 ± 0.51 | 0.20 ± 0.02 | 1.43 | |
| 3 | 0.95 ± 0.01 | 1.08 ± 0.02 | 1.92 ± 0.01 | 12.51 ± 1.1 | 7.40 ± 0.5 | 0.25 ± 0.03 | 1.27 | |
The physiological properties of each of the clones isolated from the independent endpoint ALE experiments were compared to examine whether there were any improved phenotypic outcomes across the different experiments
Fig. 3.
Growth profiles of ALE clones isolated from the end populations of the evolution experiments versus starting strains on M9 minimal medium with 20 g/L sucrose. Error bars represent standard deviation (n = 3). MG1655 construct with csc (MGcscBKA) growth profile is represented on a, MGcscBKA derivatives with cscB SNP (MGcscBKAp) mutation on b and E. coli W strain on c. Optical density measurement at OD600 were measured on a plate reader and the common ratio between the plate reader OD600 and a benchtop spectrophotometer with a 1 cm path length is 4.2
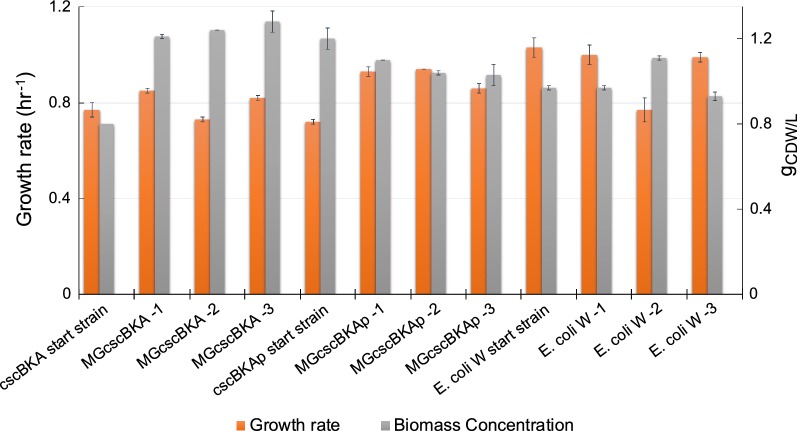
Furthermore, growth profiling of all of the sucrose-evolved endpoint clones was performed in glucose M9 media. In agreement with the observed sucrose M9 phenotypes, all clones demonstrated an increased growth rate over the start strains without any significant changes to the final cellular density (gCDW/L) (Fig. 4). One exception to this was E. coli W replicate #2 which exhibited a slightly lower growth rate than the starting strain. The observation that the vast majority of the sucrose-evolved strains also displayed a fast growth phenotype on glucose is an indication that most mutations acquired during the sucrose ALE experiment were beneficial for growth on both sugars. Such a phenotype is advantageous for a platform strain as it could be used in multiple media conditions. All isolated clones were then sequenced to examine their genetic basis along with population sequencing to better understand the genotypes responsible for the observed increases in population fitness from all of the independent ALE experiments.
Fig. 4.
Characterization of the isolated sucrose-evolved clones on 2 g/L glucose M9 minimal medium. Shown is a stacked histogram of growth rate (h−1, orange) of the selected clones and their corresponding final cellular density (gCDW/L, grey). The plot shows that mutations acquired during the ALE experiment on sucrose were also largely beneficial, to a similar extent, on glucose as a sole carbon source. Error bars represent standard deviation of biological replicates (n = 3). Data is shown for both the starting strains and evolved clones for the three different sets of evolved strains
Whole genome sequencing and mutation analysis
Whole genome sequencing was used to determine the genetic basis of the improved fitness phenotypes for the evolved E. coli strains on sucrose minimal medium. The nine isolated endpoint clones were sequenced along with the corresponding populations from which they were isolated. Overall, there were nine genes or genetic regions identified from the K-12 strains found in clones and/or populations (frequency cutoff ≥ 0.20) that were mutated either once or several times across the parallel independent replicates. This number was five for E. coli W. Additionally, there was a range of 1-3 unique mutations found in all of the clones sequenced, with most clones possessing one mutation. When looking at the population data, there was a range of 0-3 mutations detected in each sample with a frequency cutoff ≥ 0.20, indicating different levels of clonal interference in different replicates. Key mutations were identified by comparing all clones and population samples sequenced and identifying open reading frames (i.e., genes), or intragenic regions that had multiple unique mutations or were mutated across independent experiments (Table 3). Interestingly, there were no shared key mutations found between the K-12 and E. coli W experiments. Full mutation lists for each sequenced sample are given in Additional file 1: Data file S1.
Table 3.
Key mutations found after evolution of E. coli on sucrose
| Strain | Gene or genetic region | Mutation | Mutation type | Function | Number of independent occurrences | Sample ID (frequency of mutation in population samples) |
|---|---|---|---|---|---|---|
| K-12 starting strains MGcscBKA and MGcscBKAp | pyrE/rph | Δ82 bp intragenic | DEL | Orotate phosphoribosyltransferase/ribonuclease PH | 2 | MGcscBKA A1 F136 I0 (43%), MGcscBKA A1 F136 I1, MGcscBKA A3 F140 I0 (25%) |
| T→G intergenic (− 47/+ 19) | SNP | 1 | MGcscBKAp A3 F139 I0 (71%) | |||
| rpoB | S621F (TCC→TTC) | SNP | RNA polymerase subunit β | 1 | MGcscBKA A1 F136 I0 (48%), MGcscBKA A1 F136 I1 | |
| Q618L (CAG→CTG) | SNP | 1 | MGcscBKA A2 F134 I0 (4%) | |||
| rpoC | T1045P (ACC→CCC) | SNP | RNA polymerase subunit β’ | 2 | MGcscBKA A3 F140 I0 R1 (20%), MGcscBKA A3 F140 I1 R1, MGcscBKAp A3 F139 I0 (83%) | |
| R1075C (CGT→TGT) | SNP | 2 | MGcscBKAp A1 F138 I0 (93%), MGcscBKAp A1 F138 I1, MGcscBKAp A2 F140 I0 (100%), MGcscBKAp A2 F140 I1 | |||
| L770R (CTC→CGC) | SNP | 1 | MGcscBKA A3 F140 I0 (76%) | |||
| R978C (CGT→TGT) | SNP | 1 | MGcscBKAp A3 F139 I0 (9.7%), MGcscBKAp A3 F139 I1 | |||
| Q665K (CAG→AAG) | SNP | 1 | MGcscBKA A2 F134 I0 (6%) | |||
| R1174P (CGT→CCT) | SNP | 1 | MGcscBKA A2 F134 I0 (18%) | |||
| E. coli W | cscR | Δ1403 bp [cscR, dsdX, dsdA] | DEL | Sucrose operon repressor (Csc operon regulatory protein), permease DsdX, d-serine dehydratase (EC:4.3.1.18) | 1 | E. coli W A1 F155 I1 |
| Δ84 bp | DEL | Sucrose operon repressor (Csc operon regulatory protein) | 1 | E. coli W A3 F156 I1 | ||
| Δ10 bp | DEL | 1 | E. coli W A1 F155 I0 (74%) | |||
| Δ21 bp | DEL | Sucrose operon repressor (Csc operon regulatory protein) | 1 | E. coli W A3 F156 I0 (46%) | ||
| mrdB | S31R (AGC→AGG) | SNP | Rod shape-determining protein RodA | 1 | E. coli W A1 F155 I0 (77%) | |
| S37R (AGC→AGA) | SNP | 1 | E. coli W A2 F158 I0 (81) | |||
| E270Q (GAA→CAA) | SNP | 1 | E. coli W A2 F158 I1 | |||
| mrdA | W434C (TGG→TGC) | SNP | Penicillin-binding protein 2 | 1 | E. coli W A1 F155 I1 |
The sample ID has a unique identifier—(A) refers to the independent replicate, (F) refers to the flask number, and (I) to the type of the isolate: population (0) or clone (1)
The most predominantly mutated genes or intergenic regions identified across multiple independent replicates in the evolved K-12 strains were related to the RNA polymerase subunits rpoB and rpoC, as well as orotate phosphoribosyltransferase, pyrE. The first key mutations were found in the β (beta) and β’ (beta prime) subunit of RNA polymerase encoded in rpoB and rpoC, respectively [30, 31]. The number of unique mutations for each of rpoB and rpoC found across parallel populations and clones was 2 and 6 unique SNPs, respectively. Both rpoB mutations occurred in one region between amino acids residues (AAR) 618 and 621, whereas the rpoC mutations occurred closer to the carboxyl terminus (AAR total 1342) of the beta prime subunit at AAR between 665 and 1174, Table 3. There was one co-occurrence of a rpoC mutation, T1045P (ACC→CCC), which was observed in experiments starting from both K-12 strains. Interestingly, mutations in rpoB and rpoC genes have previously been found repeatedly across all different K-12 ALE experiments, indicating a very high level of parallel evolution [22, 32, 33]. It is also interesting to note that no clone contained mutations in both genes, but there were often multiple mutations in these genes found in population samples (never adding up to more than approximately 1 in mutation frequency). Specific mutations in these two subunits of the RNA polymerase, i.e. the beta and the beta prime, were found to carry beneficial growth advantages when E. coli grows on minimal medium with a range of different carbon sources [22, 32–36]. The second key mutated region observed in K-12 strains were related to pyrE expression. K-12 strains are known to possess a frame shift in rph which leads to pyrimidine starvation on minimal media and can be alleviated by mutations [37]. Similar mutations were reported in different ALE studies on different carbon sources [32, 34]. Mutations in this genetic region were found to improve fitness on minimal media with an increase in growth rate of 17% over the starting strain on glucose [22].
Strain-specific adaptive mutations in E. coli W clones were predominantly affecting a metabolic regulation pathway targeting the csc operon and cell wall biosynthesis through rod shape determining proteins. Many of the mutations found related to the csc operon were in the cscR gene, which encodes a transcriptional repressor regulator for csc operon (cscB, cscA, cscK genes) in low concentrations of sucrose [11, 16]. A cscR mutation was observed in two endpoint clones (out of three) and a total of four mutations relating to this gene were found overall when considering population sequencing. The clonal cscR mutations were an intragenic in-frame Δ84 bp deletion and an Δ1403 bp deletion which also included the dsdX and dsdA genes located next to cscR on the chromosome. Both mutations are likely a disruption of the cscR gene with potentially a loss of function. The two genes dsdX and dsdA are pseudogenes coding for a d-serine transporter and d-serine ammonia-lyase, respectively, as part of serine degradation pathway [38, 39]. Deletions of cscR were previously demonstrated to improve growth and yield for chemical bioproduction [12, 40]. Another pair of genes that were mutated several times along parallel experiments is mrdB (three times) and mrdA (one time). MrdB is annotated as rod shape-determining protein RodA [41, 42] and the related mrdA is annotated as penicillin-binding protein 2 [43]. All of the mutations found in the mrdB and mrdA genes were SNPs changing the properties of the protein through single amino acid changes (see Table 3). The impact of each SNP on the protein functionality or activity was not immediately clear based on the known structural data for these genes. Mutations in the mrdA and mrdB genes were reported in a previous temperature tolerance ALE experiment [44]. Mutations in mrd loci encoding the elongasome such as mrdA and mrdB have been shown to increase the levels of the growth-rate-regulating molecule (p)ppGpp, which potentially can lead to carbon metabolism modulation [45]. Additionally, a causal mutation in the cell shape determining gene mrdA was identified in ALE for developing osmotolerant E. coli strains [46].
Validation of mutational causality by reverse engineering
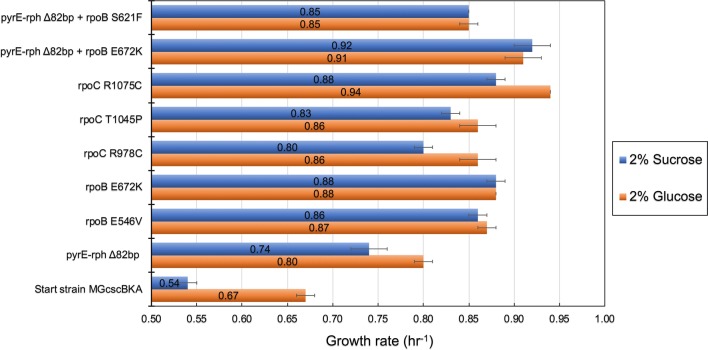
To examine the causality of key mutations identified in this study, growth screens were performed for relevant single and double mutant strains of K-12 MG1655. Such mutant strains (see methods, validation of mutation causality) were either isolated directly from the current study (i.e., endpoint clones containing the csc construct and mutations) or generated from previously constructed mutant strains [22] which were subsequently engineered to also contain the csc gene cassette (see Table 4). Growth screens were performed side by side on M9 medium supplemented with either sucrose or glucose (20 g/L). Genes investigated for potential causality for improved fitness, i.e., higher growth rate, were rpoC, rpoB and the intergenic deletion ∆82 bp between pyrE/rph. To put these results in context, ALE-derived mutations in these exact genes were validated previously to confer fitness advantages in various substrate environments, and in particular when grown on glucose [22, 32, 33]. Figure 5 provides comparative fitness levels for each adaptive mutation on either sucrose or glucose.
Table 4.
Source of key mutations validated for causality in isolated strains from the current study or constructed previously from Lacroix et al. [22]
| Genetic region | Mutation | Source |
|---|---|---|
| K-12 MG1655 | N/A | N/A |
| pyrE-rph | Δ82 bp deletion | LaCroix et al. [22] |
| rpoB | E672K (GAA→AAA) | LaCroix et al. [22] |
| rpoC | R1075C (CGT→TGT) |
MGcscBKAp A1 F138 I1 MGcscBKAp A2 F140 I1 |
| R978C (CGT→TGT) | MGcscBKAp A3 F139 I1 | |
| T1045P (ACC→CCC) | MGcscBKA A3 F140 I1 with additional mutations of (tdcG, baeS) | |
| pyrE-rph + rpoB | Δ82 bp + E672K (GAA→AAA) | LaCroix et al. [22] |
| pyrE-rph + rpoB | Δ82 bp + S621F (TCC→TTC) | MGcscBKA A1 F136 I1 |
Fig. 5.
Causal mutation analysis for fitness improvement on sucrose and glucose for K-12 strains. Shown is a bar graph representing growth rate of identified causal mutations from the current study (ALE on sucrose) and LaCroix et al. [22] (ALE on glucose). The error bars are calculated from two biological replicates (n = 2). All rpoC mutations originated from this study, as well as the rpoB S621F with the pyrE-rph deletion mutation (i.e., they were not reverse engineered). The remaining mutations were from glucose ALE control experiment (GLUALE) with chromosomally integrated csc gene cassette as described in the methods
The comparative analysis of reverse engineered strains showed that mutated genes identified in this ALE study conferred a fitness advantage over the starting strain and some were additive in their impact on growth rate. Interestingly, the fitness advantage of all tested mutations over the starting strain was observed when grown on M9 with either sucrose or glucose as carbon sources, and the fitness level increases for a given mutated strain were roughly similar when grown on sucrose or glucose. For the mutations examined, the RNA polymerase mutations have a higher increase in growth rate over the causal mutation affecting nucleotide biosynthesis (pyrE). Furthermore, analysis showed that when multiple mutations were present in a strain, such double mutant strains conferred a higher fitness advantage over single mutants with the identical mutations. Specifically, a strain with a mutation in the intragenic region between pyrE/rph genes and in rpoB (E672K) had higher fitness than strains with either single mutations. This pattern of co-occurrence was observed when evolving K-12 MG1655 on either glucose [22] or sucrose (endpoint isolate derived from MGcscBKA ALE#1 population from the current study).
Benchmarking of evolved strains performance on sugarcane juice
To further evaluate the ALE-derived clones for sucrose utilization, each endpoint clone was characterized on a raw feedstock, sugarcane juice (SCJ) medium. A M9 medium base was utilized with SCJ to test both evolved and starting strains (Table 2 and Additional file 2: Figs. S1–S3). Raw SCJ mainly consists of three carbon sources; sucrose with the highest fraction, and glucose and fructose in small fractions [47]. The sucrose uptake rates (SURs) and acetate production rates (APRs) for all of the isolated clones were compared to each of the starting strains. Overall, there was an increase in the SUR for the K-12 MG1655 and E. coli W endpoint clones. The average increase in SUR was 1.10- ± 0.05- and 1.35 ± 0.05-fold increase for MGcscBKA and MGcscBKAp isolates, respectively, and a round 1.2 ± 0.2-fold increase for E. coli W isolates. Analysis of the MGcscBKAp starting strain showed a slightly lower SUR than that of the MGcscBKA starting strain. This finding was in contrast to the observation that such a mutation in cscB leads to a two-fold increase in sucrose uptake rate in a different E. coli strain [16]. However, the evolved MGcscBKAp clones demonstrated higher uptake rates than the MGcscBKA evolved clones (Table 2). Thus, it appears that there was an impact from this mutation at faster growth rates displayed by the evolved clones, but not the starting strains. However, reverse engineering of this mutation in the mutated endpoint strains of the MGcscBKA evolved clones would be needed to show this definitively.
As a general observation on the growth profiles of the evolved isolates, the initial amounts of glucose and fructose (approximately, 1–2 mM) were first depleted followed by the consumption of sucrose (Additional file 2: Figs. S1–S3). Interestingly, during the exponential growth phase, fructose and glucose accumulated in the culture broth in some replicates (e.g., E. coli W endpoint replicate #1, Additional file 2: Fig. S1) and was then consumed near the end of the aerobic cultivation. It was also observed that acetate was produced and accumulated during the course of the cultivation for all the tested clones except for the E. coli W clones (Additional file 2: Figs. S1–S3). In the evolved W clones, acetate accumulated up until approximately 10 h after inoculation, and then it was consumed as the sucrose, glucose, and fructose were depleted.
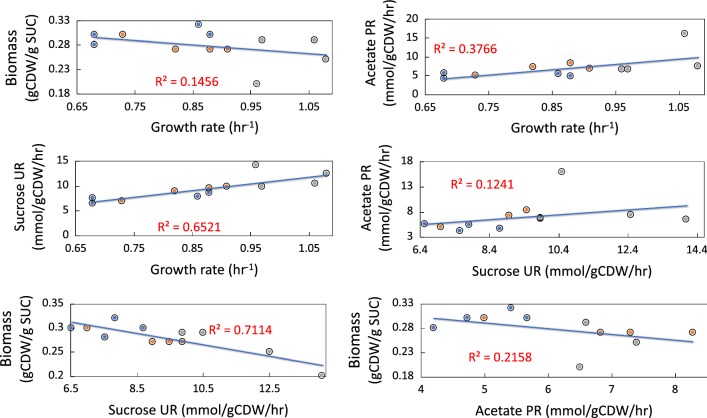
The physiological profiling of selected endpoint isolate on SCJ were compared to examine whether there were any correlations or observed trends across the different evolved replicate experiments. Although the evolved clones isolated from the endpoints of the ALE experiments mostly showed a similar increase in fitness, the SUR and biomass yields varied more significantly (Table 2). However, the SURs and APRs were most often higher in endpoint clones compared to the starting strains (the exceptions being one clone, MGcscBKA isolate #2, in which SUR was not significant). There was a weak correlation (r2 = 0.12) between the increase observed in both SUR and APR, Fig. 6. Alternatively, there was a stronger correlation between the increase in SURs and APRs and the increased growth rate with correlations of (r2 = 0.65) and (r2 = 0.37), respectively. For the characterized clones, a trade-off between the SURs and biomass yield at the steady state (YX/S) was observed where higher SURs led to lower biomass yields, i.e. an inverse correlation with an r2 = 0.71. The results implied that there were multiple mutational mechanisms through which a similar increase in the growth rate could be achieved (i.e., alternative optimal solution). The observed variations in SURs, APRs, and biomass yields further showed that under the selection pressure applied via ALE, the populations adapted through different trajectories in order to reach an apparent optimum metabolic state enabling a fast growth phenotype on sugarcane juice.
Fig. 6.
Pairwise comparisons of the phenotypic properties of the evolved clones on sugarcane juice M9 minimal medium (SCJ medium). Blue filled circles correspond to MGcscBKA clones, orange filled circles to MGcscBKAp clones and grey filled circles to E. coli W. Overall, sucrose uptake rate versus biomass yield and growth rate versus sucrose uptake rate were the highly correlated. Calculations can be seen in the methods section
Discussion
Platform strains that are able to exploit sucrose as abundant cheap carbon source from raw materials sources such as molasses and sugarcane juice can contribute to the increased economic and environmental sustainability of bio-based chemical production. Most of previous attempts to engineer a fast-growing K-12 strain on sucrose, to the same rate as growth on glucose, have not been successful. Thus, the scope of the current study was to generate a platform strains able to utilize sucrose as a carbon source and grow rapidly using adaptive laboratory evolution. Accordingly, the main findings from the current work are: (1) it was possible to effectively generate platform strains with elevated growth rates in a sucrose feedstock environment, which includes validation on raw material sugarcane juice. Moreover, the developed platform strains are suitable as fermentation chassis with an extended capability to also rapidly utilize glucose to the same level as strains evolved to grow solely on glucose [22]; (2) this study establishes a transferable sucrose utilization cassette for use in K-12 strains where sucrose utilization is not native; and (3) the identification of key reproducibly-occurring mutations using multiple parallel replicate ALE experiments. These mutations were validated using knock-in strains and can be used as cell engineering parts in additional strains of interest.
The ALE approach utilized in this work was successful in generating strains with improved fitness over their respective starting strains after approximately 40 days of continuous passaging in the exponential growth phase. The selected clones derived from the evolved populations for the K-12 strains exhibited increased carbon uptake rates for both sucrose and glucose, making them suitable for both carbon sources. Furthermore, the approach utilized in this work to chromosomally integrate the csc cassette was successful in generating a genetically stable strain (see Additional file 2: Text), as demonstrated throughout the extent of work in the study. This is in contrast to previously published plasmid expressions that have been shown to lead to complications in industrial production due to requirements for the addition of antibiotics and the burden of maintaining high-copy numbers plasmids [15, 48].
The key mutations identified in this study can be used as engineering parts for strain engineering. Given that a similar set of mutations enables enhanced growth on both glucose and sucrose, it is reasonable to expect that such mutations and strains have growth benefits on similar substrates, or mixes of such substrates, and this represents an avenue forward for generating a more universally applicable platform strain [34, 36, 49]. Previously, the key mutations identified in the RNA polymerase subunits (RNAP) rpoB and rpoC were found to have large-scale systematic transcriptional changes that influence specific cellular processes and to be responsible for fast growth [34, 36] on minimal medium. The benefits of such mutations in bioprocessing have been demonstrated and establish how strains with such mutations can serve as platform strains for enhanced production phenotypes [50]. Consequently, it appears that the fast-growing phenotype of K-12 on sucrose is more likely more due to a systems level regulatory change rather than changes specific to sucrose utilization.
There are a number of avenues to explore to build off of the results from this study. Such work could include an effort to study the impact of the key mutations individually, or in combination, on the fast growth phenotype. One interesting feature to pursue was that the evolved K-12 strains demonstrated lower SURs on sucrose than the W strain and this could be further examined. It has been shown that E. coli W maintains a highly oxidative metabolic state either on glucose or sucrose with low accumulation of overflow metabolites such as acetate [51]. The two E. coli strains possess significantly different genome sizes (4.90 Mbp for W vs 4.64 Mbp for K-12) with different numbers of genes, pseudogenes, and mobile elements [10], therefore a comparative genomic study to associate this particular feature was not apparent. A potentially fruitful approach to address this observed difference would likely include growth of the evolved and unevolved E. coli W and K-12 strains along with multiple omics assays to better understand the underlying mechanisms, as performed previously [36], in a sucrose utilizing context. Further, such an analysis with an engineered production pathway would also place the evolved phenotypes in a relevant production context.
In summary, ALE has been demonstrated to generate strains with reproducible causal mutations for increased sucrose utilization for E. coli under a strict growth rate selection pressure. Whole genome re-sequencing revealed the underlying causal mutations of the evolved strains. Thus, the ALE derived clones and the beneficial mutations represent a promising platform for developing a sucrose-based bioproduction chassis and can provide a good starting point to develop sucrose utilization in other industrially relevant E. coli strains.
Additional files
Additional file 1: Data file S1. All mutation list of ALE resequenced clones and populations.
Additional file 2: Text file S1. Full strains design description and different genetic manipulation methods.
Acknowledgements
We would like to thank Mohammad Radi, Patrick Phaneuf and Anna Koza for their help in the project. Funding for the project was provided by The Novo Nordisk Foundation Grant Numbers NNF14OC0011269 and NNF10CC1016517. We greatly appreciate the help of Tim Mies (Energy Biosciences Institute, University of Illinois for collecting and storing the cane juice utilized in this study. This part of the research was supported by the Energy Biosciences Institute (project OO2J14).
Abbreviations
- ALE
adaptive laboratory evolution
- SUC
sucrose
- SUR
sucrose uptake rate
- APR
acetate production rate
- GLUALE
glucose adaptive laboratory evolution
- RNAP
RNA polymerase
- AAR
amino acids residues
Authors’ contributions
Conceived and designed the experiments: ETM, AMF. Performed most of the experiments; ETM. JL constructed the pOSIP-csc plasmids and JL, ETM used it for chromosomal integration of the csc cassette. Contributed materials, i.e. Sugarcane Juice: IC and RIM. Wrote the paper: ETM and AMF. HM, JL, MJH, ATN, IC and RIM; reviewed manuscript and helped in discussing the results. All authors read and approved the final manuscript.
Funding
Funding for the project was provided by The Novo Nordisk Foundation Grant Numbers NNF14OC0011269 and NNF10CC1016517.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its additional files].
Ethics approval and consent to participate
Not applicable.
Consent of publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elsayed T. Mohamed, Email: ettm@biosustain.dtu.dk
Hemanshu Mundhada, Email: hmun@biosustain.dtu.dk.
Jenny Landberg, Email: jenla@biosustain.dtu.dk.
Isaac Cann, Email: icann@illinois.edu.
Roderick I. Mackie, Email: r-mackie@illinois.edu
Alex Toftgaard Nielsen, Email: atn@biosustain.dtu.dk.
Markus J. Herrgård, Email: herrgard@biosustain.dtu.dk
Adam M. Feist, Email: afeist@ucsd.edu
References
- 1.Bevan MW, Franssen MCR. Investing in green and white biotech. Nat Biotechnol. 2006;24:765. doi: 10.1038/nbt0706-765. [DOI] [PubMed] [Google Scholar]
- 2.Koutinas AA, Wang R, Webb C. Evaluation of wheat as generic feedstock for chemical production. Ind Crops Prod. 2004;20:75–88. doi: 10.1016/j.indcrop.2003.12.013. [DOI] [Google Scholar]
- 3.Renouf MAA, Wegener MKK, Nielsen LKK. An environmental life cycle assessment comparing Australian sugarcane with US corn and UK sugar beet as producers of sugars for fermentation. Biomass Bioenerg. 2008;32:1144–1155. doi: 10.1016/j.biombioe.2008.02.012. [DOI] [Google Scholar]
- 4.Fadel M, Keera AA, Mouafi FE, Kahil T. High level ethanol from sugar cane molasses by a new thermotolerant Saccharomyces cerevisiae strain in industrial scale. Biotechnol Res Int. 2013;2013:253286. doi: 10.1155/2013/253286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung MY, Jung HM, Lee J, Oh MK. Alleviation of carbon catabolite repression in Enterobacter aerogenes for efficient utilization of sugarcane molasses for 2,3-butanediol production. Biotechnol Biofuels. 2015;8:106. doi: 10.1186/s13068-015-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monk JMM, Koza A, Campodonico MAA, Machado D, Seoane JMM, Palsson BOO, et al. Multi-omics quantification of species variation of Escherichia coli links molecular features with strain phenotypes. Cell Syst. 2016;3(238–251):e12. doi: 10.1016/j.cels.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baeshen MN, Al-Hejin AM, Bora RS, Ahmed MMM, Ramadan HAI, Saini KS, et al. Production of biopharmaceuticals in E. coli: current scenario and future perspectives. J Microbiol Biotechnol. 2015;25:953–962. doi: 10.4014/jmb.1412.12079. [DOI] [PubMed] [Google Scholar]
- 8.Idalia V-MN, Bernardo F. Escherichia coli as a model organism and its application in biotechnology. Recent Adv Physiol Pathog Biotechnol Appl. 2017. 10.5772/67306.
- 9.Bergey DH, David H, Holt JG. Bergey’s manual of determinative bacteriology. Medical; 1994. https://books.google.se/books/about/Bergey_s_Manual_of_Determinative_Bacteri.html?id=jtMLzaa5ONcC&redir_esc=y. Accessed 4 Jun 2019.
- 10.Archer CT, Kim JF, Jeong H, Park JH, Vickers CE, Lee SY, et al. The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli. BMC Genomics. 2011;12:9. doi: 10.1186/1471-2164-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bockmann J, Heuel H, Lengeler JW. Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Mol Gen Genet. 1992;235:22–32. doi: 10.1007/BF00286177. [DOI] [PubMed] [Google Scholar]
- 12.Sabri S, Nielsen LK, Vickers CE. Molecular control of sucrose utilization in Escherichia coli W, an efficient sucrose-utilizing strain. Appl Environ Microbiol. 2013;79:478–487. doi: 10.1128/AEM.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L-CC, Hor L-II, Wu J-YY, Chen T-LL. Effect of specific growth rate on the production of a recombinant nuclease by Escherichia coli. Biochem Eng J. 2003;14:101–107. doi: 10.1016/S1369-703X(02)00156-0. [DOI] [Google Scholar]
- 14.Zawada J, Swartz J. Maintaining rapid growth in moderate-density Escherichia coli fermentations. Biotechnol Bioeng. 2005;89:407–415. doi: 10.1002/bit.20369. [DOI] [PubMed] [Google Scholar]
- 15.Diaz Ricci JC, Hernández ME. Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol. 2000 doi: 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- 16.Jahreis K, Bentler L, Bockmann J, Hans S, Meyer A, Siepelmeyer J, et al. Adaptation of sucrose metabolism in the Escherichia coli wild-type strain EC3132. J Bacteriol. 2002;184:5307–5316. doi: 10.1128/JB.184.19.5307-5316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JW, Choi S, Park JH, Vickers CE, Nielsen LK, Lee SY. Development of sucrose-utilizing Escherichia coli K-12 strain by cloning β-fructofuranosidases and its application for l-threonine production. Appl Microbiol Biotechnol. 2010;88:905–913. doi: 10.1007/s00253-010-2825-7. [DOI] [PubMed] [Google Scholar]
- 18.Shukla VB, Zhou S, Yomano LP, Shanmugam KT, Preston JF, Ingram LO. Production of l(−)-lactate from sucrose and molasses. Biotechnol Lett. 2004;26:689–693. doi: 10.1023/B:BILE.0000024088.36803.4e. [DOI] [PubMed] [Google Scholar]
- 19.Tsunekawa H, Azuma S, Okabe M, Okamoto R, Aiba S. Acquisition of a sucrose utilization system in Escherichia coli K-12 derivatives and its application to industry. Appl Environ Microbiol. 1992;58:2081–2088. doi: 10.1128/aem.58.6.2081-2088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruschi M, Boyes SJ, Sugiarto H, Nielsen LK, Vickers CE. A transferable sucrose utilization approach for non-sucrose-utilizing Escherichia coli strains. Biotechnol Adv. 2012;30:1001–1010. doi: 10.1016/j.biotechadv.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Lee SY, Kim HU. Systems strategies for developing industrial microbial strains. Nat Biotechnol. 2015;33:1061–1072. doi: 10.1038/nbt.3365. [DOI] [PubMed] [Google Scholar]
- 22.LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, et al. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, et al. Evolution of Escherichia coli to 42 C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol. 2014;31:2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St-Pierre F, Cui L, Priest DG, Endy D, Dodd IB, Shearwin KE. One-step cloning and chromosomal integration of DNA. ACS Synth Biol. 2013;2:537–541. doi: 10.1021/sb400021j. [DOI] [PubMed] [Google Scholar]
- 25.Cui Lun, Shearwin Keith E. Methods in Molecular Biology. New York, NY: Springer New York; 2016. Clonetegration Using OSIP Plasmids: One-Step DNA Assembly and Site-Specific Genomic Integration in Bacteria; pp. 139–155. [DOI] [PubMed] [Google Scholar]
- 26.Phaneuf PV, Gosting D, Palsson BO, Feist AM. ALEdb 1.0: a database of mutations from adaptive laboratory evolution. bioRxiv. 2018;320747. 10.1101/320747v1.full. [DOI] [PMC free article] [PubMed]
- 27.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaCroix RA, Palsson BO, Feist AM. A model for designing adaptive laboratory evolution experiments. Appl Environ Microbiol. 2017 doi: 10.1128/aem.03115-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee DH, Feist AM, Barrett CL, Palsson B. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli. PLoS ONE. 2011;6:e26172. doi: 10.1371/journal.pone.0026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross W, Gosink K, Salomon J, Igarashi K, Zou C, Ishihama A, et al. A third recognition element in bacterial promoters: DNA binding by the subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 31.Simpson RB. The molecular topography of rna polymerase-promoter interaction. Cell. 1979;18:277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- 32.Sandberg TE, Lloyd CJ, Palsson BO, Feist AM. Laboratory evolution to alternating substrate environments yields distinct phenotypic and genetic adaptive strategies. Appl Environ Microbiol. 2017 doi: 10.1128/aem.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR. Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab Eng. 2017;44:100–107. doi: 10.1016/j.ymben.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conrad TM, Frazier M, Joyce AR, Cho B-K, Knight EM, Lewis NE, et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohamed ET, Wang S, Lennen RM, Herrgård MJ, Simmons BA, Singer SW, et al. Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution. Microb Cell Fact. 2017;16:1–15. doi: 10.1186/s12934-017-0819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Utrilla J, O’Brien EJ, Chen K, McCloskey D, Cheung J, Wang H, et al. Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution. Cell Syst. 2016;2:260–271. doi: 10.1016/j.cels.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norregaard-Madsen M, McFall E, Valentin-Hansen P. Organization and transcriptional regulation of the Escherichia coli K-12 d-serine tolerance locus. J Bacteriol. 1995;177:6456–6461. doi: 10.1128/jb.177.22.6456-6461.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arifin Y, Sabri S, Sugiarto H, Krömer JO, Vickers CE, Nielsen LK. Deletion of cscR in Escherichia coli W improves growth and poly-3-hydroxybutyrate (PHB) production from sucrose in fed batch culture. J Biotechnol. 2011;156:275–278. doi: 10.1016/j.jbiotec.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, Suh H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016;1:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishino F, Park W, Tomioka S, Tamaki S, Takase I, Kunugita K, et al. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J Biol Chem. 1986;261:7024–7031. [PubMed] [Google Scholar]
- 44.Deatherage DE, Kepner JL, Bennett AF, Lenski RE, Barrick JE. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc Natl Acad Sci. 2017;114:E1904–E1912. doi: 10.1073/pnas.1616132114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sperber AM, Herman JK. Metabolism shapes the cell. J Bacteriol. 2017;199:1–14. doi: 10.1128/JB.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winkler JD, Garcia C, Olson M, Callaway E, Kao KC. Evolved osmotolerant escherichia coli mutants frequently exhibit defective N-acetylglucosamine catabolism and point mutations in cell shape-regulating protein MreB. Appl Environ Microbiol. 2014;80:3729–3740. doi: 10.1128/AEM.00499-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Godshall MA, Legendre BL. SUGAR|Sugarcane. Encycl Food Sci Nutr. 2003;56:45–51. [Google Scholar]
- 48.Wang Z, Xiang L, Shao J, Wegrzyn A, Wegrzyn G. Effects of the presence of CoIE1 plasmid DNA in Escherichia coli on the host cell metabolism. Microb Cell Fact. 2006;5:1–18. doi: 10.1186/1475-2859-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rugbjerg P, Feist AM, Sommer MOA. Enhanced metabolite productivity of Escherichia coli adapted to glucose m9 minimal medium. Front Bioeng Biotechnol. 2018;6:166. doi: 10.3389/fbioe.2018.00166/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arifin Y, Archer C, Lim SA, Quek LEE, Sugiarto H, Marcellin E, et al. Escherichia coli W shows fast, highly oxidative sucrose metabolism and low acetate formation. Appl Microbiol. 2014;98:9033–9044. doi: 10.1007/s00253-014-5956-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Data file S1. All mutation list of ALE resequenced clones and populations.
Additional file 2: Text file S1. Full strains design description and different genetic manipulation methods.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its additional files].