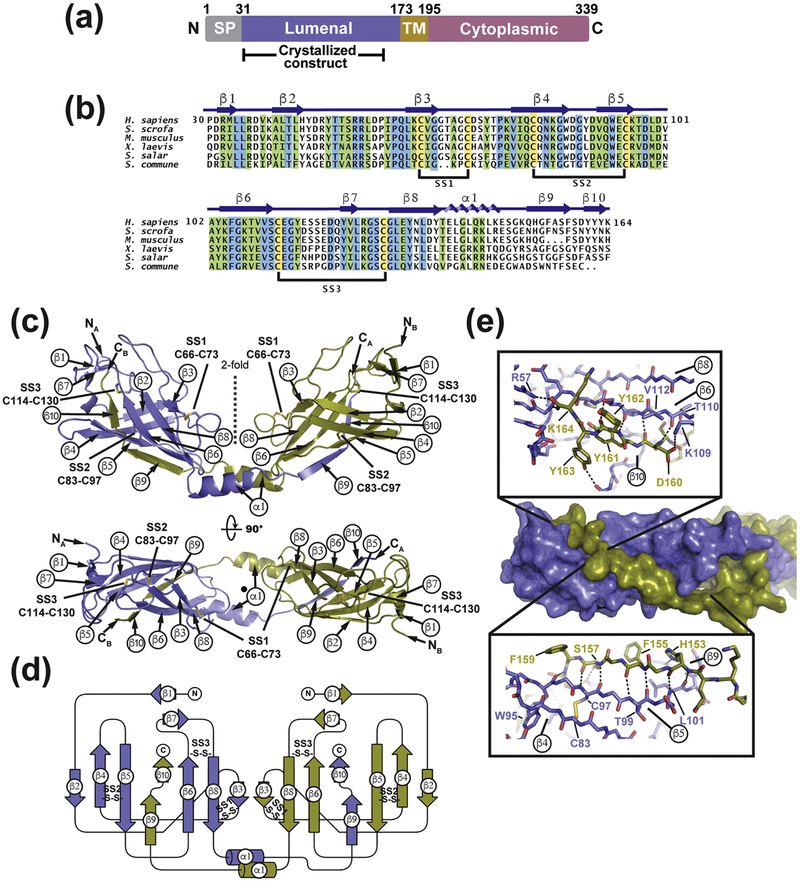
Fig. 1. Structure of SARAFL.
(a) SARAF schematic. Signal peptide (SP, grey), luminal (purple), transmembrane (TM, brown), and cytoplasmic (magenta) domains and amino acid boundaries of each are indicated. Extent of crystallized SARAFL construct is shown. (b) SARAFL sequence comparison from human (H. sapiens), pig (S. scrofa), mouse (M. musculus), frog (X. laevis), salmon (S. salar), and mushroom (S. commune). Invariant (blue), conserved (green) and cysteines (yellow) are highlighted. Secondary structure elements and disulfide bonds (SS1, SS2, and SS3) are shown. (c) Structure of the SARAFL domain swapped dimer. N and C termini (NA, NB, CA, and CB) and secondary structure elements of each chain are labeled. Disulfide bonds are shown as sticks and are labeled. Chains A and B are slate and deep olive, respectively. (d) SARAFL domain swapped dimer topology diagram. ‘S-S’ denotes disulfide bonds. (e) Detailed view of interactions of domain swapped β 9 and β 10 with the SARAFL core. Dashed lines indicate hydrogen bonds.