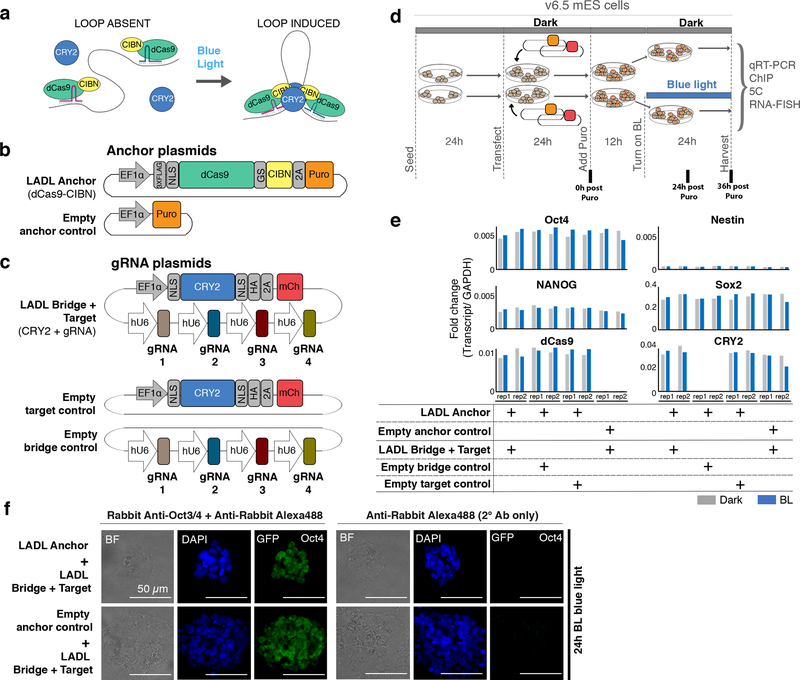
Figure 1. Concept, design, and implementation of the light-activated dynamic looping (LADL) system.
(a) Schematic of the LADL system. In the dark, the LADL anchoring protein dCas9-CIBN is recruited to two specific genomic fragments using guide RNAs (gRNAs). CRY2 is soluble and does not specifically associate with chromatin. Upon blue light illumination, CRY2 homodimerizes and CRY2 and dCas9-CIBN heterodimerize to form a bridge connecting the two gRNA-bound genomic fragments. We hypothesize that the bridge would loop out the intervening DNA. (b-c) Schematic of plasmid constructs encoding the (b) puromycin-selectable LADL Anchor and Empty anchor control and (c) the LADL Bridge + Target, the Empty target control, and the Empty bridge control. (d) Schematic timeline of seeding, transfection, puromycin selection, and blue light illumination of v6.5 mouse embryonic stem cells. (e) qRT-PCR analysis of Oct4, Nestin, Nanog, Sox2, dCas9, and CRY2 transcript levels in co-transfected mouse embryonic stem cells after 36 hours post-puromycin selection. Data from two independent experiments are shown. (f) Immunofluorescence staining for Oct4 in mouse embryonic stem cells co-transfected with the indicated plasmids. Scale bars, 50 μm. Images are representative of 3 independent experiments.