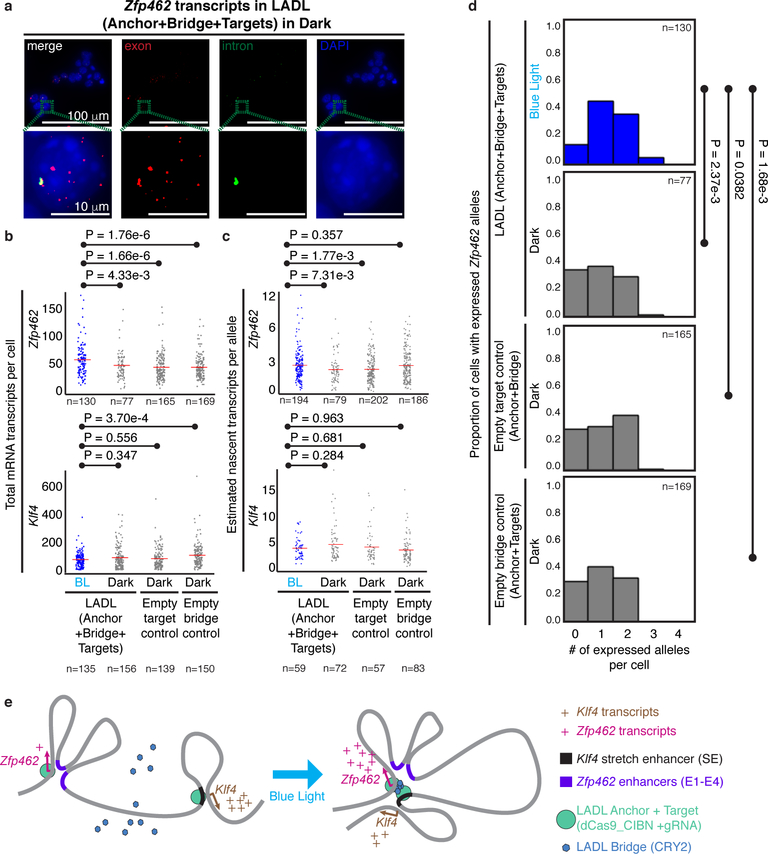
Figure 5. Functional effect of the LADL-engineered de novo loop on endogenous gene expression.
(a) Full field (top row) and zoomed microscopy images (bottom row) from single molecule RNA fluorescence in situ hybridization (FISH) analysis of LADL-engineered mouse embryonic stem cells in dark. Quantitative image analysis of Zfp462 and Klf4 transcripts from LADL engineered mouse ES cells after 24 hours of blue light (detailed in Supplementary Methods). Scale bars, 100 μm. Images are representative of n=3 independent experiments. (b-c) Strip charts representing (b) the total number of mRNA transcripts per cell and (c) the estimated number of nascent transcripts per allele for Zfp462 (upper row) and Klf4 (lower row). Red bars, means of each condition. (d) Histograms represent the proportion of cells with a specific number of actively expressing Zfp462 alleles. P-values computed using the unpaired one-tailed Mann Whitney U test. Sample sizes represent (b, d) number of cells or (c) number of active transcription alleles. Additional replicates from independent experiments are shown in Supplementary Figure 13. (e) Schematic diagram illustrating the link between LADL engineered de novo loop formation and changes in endogenous gene expression.