Fig. 1.
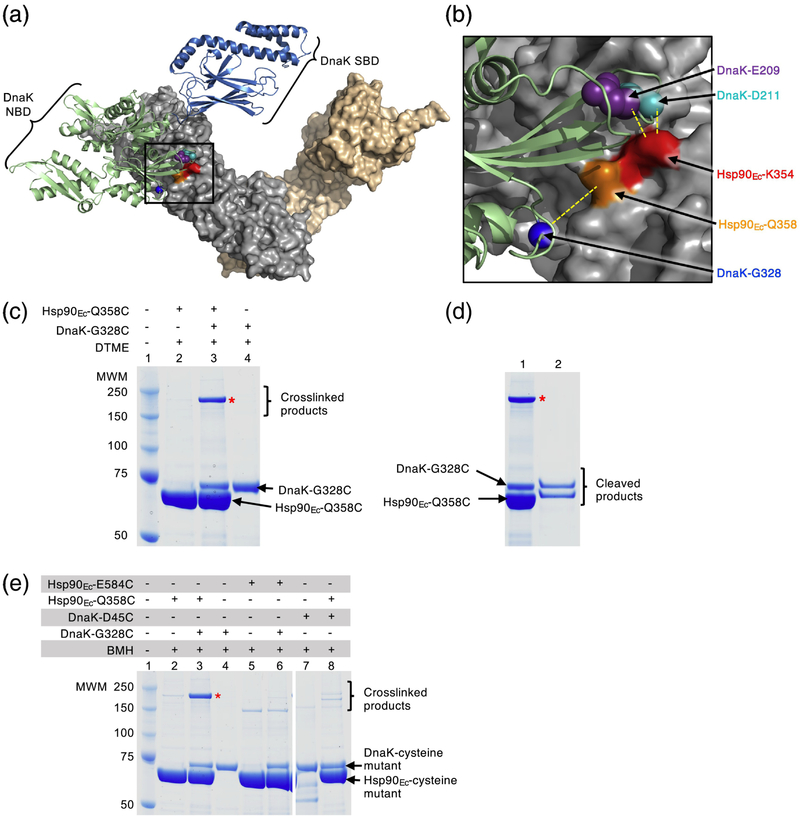
Hsp90Ec and DnaK residues directly interact. (a) Docked model [32] of the apo structure of Hsp90Ec (PDB ID code 2IOQ) [15] and ADP-bound DnaK (PDB ID code 2KHO) [73]. The apo conformation of the Hsp90Ec dimer is shown as a surface rendering with one protomer in gray and one protomer in wheat. DnaK in the ADP-bound conformation [73] is shown as a ribbon model with the NBD in light green and the SBD in blue. Residues in Hsp90Ec, K354 and Q358, that were mutated in this study are shown in red and orange, respectively, while residues in DnaK, E209, D211 and G328, that were mutated in this study are shown as CPK models in purple, cyan and blue, respectively. The black square represents the field enlarged in (b). (b) Enlargement of the interaction region between Hsp90Ec and DnaK shown in (a). Hsp90Ec and DnaK are colored as in (a) and residues mutated in this study are also as in (a). Yellow dashed lines indicate the pairs of Hsp90Ec and DnaK residues that were used in crosslinking studies. (a) and (b) were both rendered using PyMOL (https://pymol.org/2/). (c) Hsp90Ec-Q358C and DnaK-G328C (4 μM each) were treated with the crosslinker DTME (13.3 Å linker) alone and in a mixture together and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. (d) The crosslinked product from (c) lane 3 was extracted and treated with a reducing agent and the products analyzed by SDS-PAGE followed by Coomassie blue staining. (e) Hsp90Ec mutants Hsp90Ec-Q358C and E584C (4 μM), and DnaK mutants DnaK-G328C and D45C (4 μM), were treated with BMH (13 Å linker) alone and in mixtures as indicated and covalent bond formation was monitored by SDS-PAGE followed by Coomassie blue staining. Higher molecular weight bands in lanes 2, 5, 6 and 8 are likely crosslinked dimers of Hsp90Ec-Q358C (lane 2 and 8) and E584C (lane 5 and 6); the potential Hsp90Ec-E584C dimer appears to run at a lower molecular weight than other crosslinked products likely due to it attaining a more compact conformation following crosslinking. In (c-e), (*) indicates crosslinked product of interest and the gels shown are representative of at least three independent experiments.