Extended Data Figure 1. Defective nuclear envelope assembly on lagging chromosomes (or chromosome bridges) generated by multiple methods.
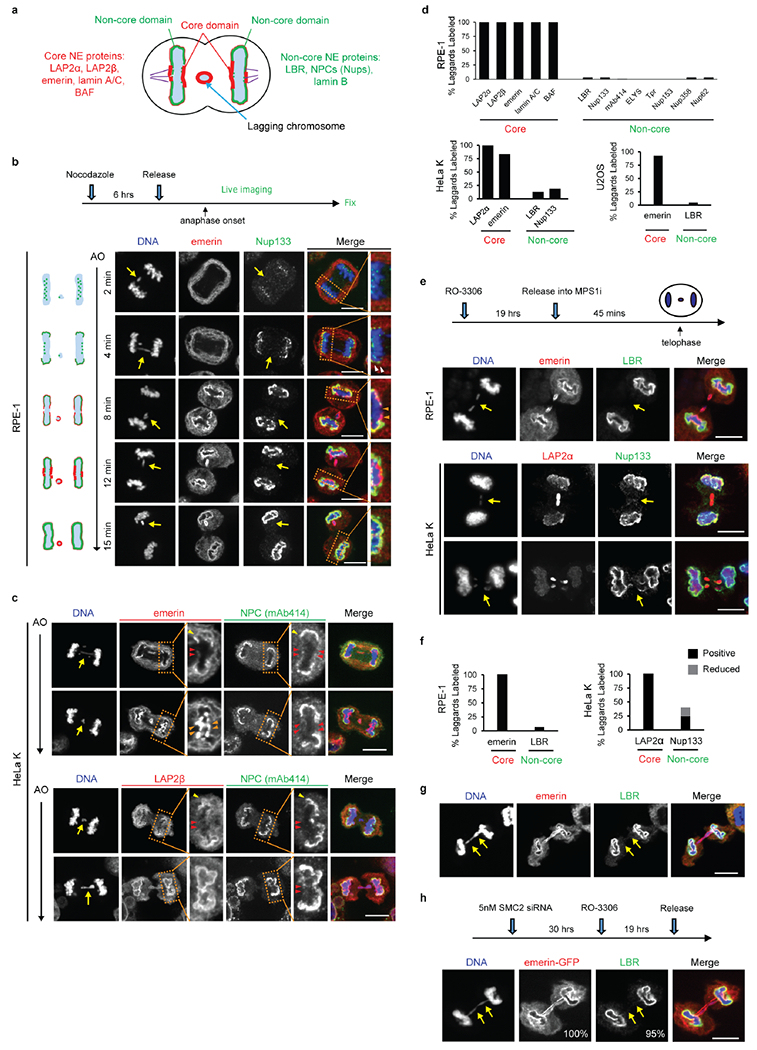
a, Cartoon showing the NE subdomains that transiently form around the main chromosome mass in a telophase cell8,9 and summarizing results reported here for lagging chromosomes. DNA is shown in light blue. Lamina-associated polypeptide 2α/β (LAP2α/β). Barrier-to-autointegration factor (BAF). Lamin B receptor (LBR). The NPC is a complex containing nucleoporins (Nups), including Nup133, Nup107, ELYS, Tpr, Nup153, Nup358 and Nup621 (see Extended Data Figs 1d, 2f).
b, Live-cell/ fixed cell imaging showing the recruitment pattern of core (emerin) and non-core (Nup133) proteins onto lagging chromosomes (yellow arrows) and the main chromosome mass at the indicated timepoints after anaphase onset (AO, t=0). Top: Experimental scheme. RFP-H2B-expressing RPE-1 cells were released from the nocodazole mitotic block, imaged on gridded coverslips at 2 min intervals, and then fixed and labeled for immunofluorescence (as in Extended Data Fig. 4e). Bottom: cartoons (left) and images of RPE-1 cells showing DNA (blue), emerin (red) and Nup133 (green). Each image is representative of 20 cells (from 2 technical replicates) from the indicated timepoints. Scale bars, 10 μm. Note: there is some variability between cells (~1–2 min) for the timing at which different proteins are recruited. During early-mid anaphase (2–4 min post AO), Nup133 is on kinetochores as previously described35 (green dots in the cartoon). ~4–6 min post AO, Nup133 begins to assemble on the chromosome periphery (enlarged image in the right most column, white arrowheads). ~6–8 min post AO, the membrane protein emerin assembles on lagging chromosome and the main chromosome mass, including the region adjacent to the central spindle (enlarged image in the right most column, orange arrowheads, microtubules are not shown). By 8–12 min post AO, emerin becomes concentrated in a recognizable “core” domain8, which is also detected as a gap in Nup133 signal (enlarged image). The peripheral localization of Nup133 (e.g., 8 min) marks the “non-core” domain, cartooned in (a). About half of the NPCs of the interphase nucleus assemble in this ~ 8–10 minute period of telophase28 (hereafter referred to as late mitotic NPC assembly). Note that nuclear pore proteins display the most obvious “non-core” gap, whereas other non-core proteins, such as LBR, more commonly display reduced signal intensity within the core domain (Extended Data Fig. 1e, top row, RPE-1), rather than absence of signal in this region. Defective Nup133 assembly on lagging chromosomes persists throughout the mitotic exit (>15 min post AO). By ~15 min post AO, the core and non-core domains become intermingled on the main nucleus, with fragments of the core domain persisting as “pore-free” islands that slowly populated by NPCs during interphase10,36.
c, Similar to RPE-1 cells (see b above), images of HeLa Kyoto (HeLa K) cells (representative of 30 cells, from 2 experiments) showing the “core” membrane proteins emerin (top two rows) and LAP2β (bottom two rows) first associate with the chromosome periphery (yellow arrowheads) contemporaneously with the non-core (NPC, mAb414 detects FG-containing nucleoporins.) proteins. ~2–4 min later, the core proteins extend into and then concentrate (emerin, orange arrowheads; LAP2β does not concentrate) in the core domain (red arrowheads). Cells were synchronized as in Extended Data Fig. 1e. Note in HeLa K cells, lagging chromosomes often exhibit a slight delay (~1–2 min) in the recruitment of core membrane proteins (emerin and LAP2β) as compared to the periphery of the main chromosome mass. Scale bars, 10 μm.
Discussion: It is thought that the NE likely assembles from a continuous network of mitotic endoplasmic reticulum (ER)1. It is therefore simplest to propose that the core and non-core subdomains are also in a continuous network, but the core domain is just a region of the continuous ER network that is missing the non-core subgroup of proteins. Supporting this idea, prior work8 and our data (b, c above) show that the core proteins initially assemble together with the non-core proteins around the chromosome periphery and only later become enriched in the regions near the microtubules. This data suggests a model that the domain partitioning could come solely from NPC precursors and LBR (which requires ELYS for recruitment37,38) being preferentially retained in fenestrated ER sheets28 that might less readily penetrate bundled spindle microtubules (see Fig. 3a). Although we favor this model, at this point, we cannot exclude the alternative that core and non-core proteins somehow partition into separate membrane compartments.
d, Quantitation of defective non-core NE protein recruitment to lagging chromosomes in different cell lines. Synchronization as in Fig. 1a (n=64, 118, 151, 124, 151, 145, 150, 69, 90, 65, 70, 60, 64, left to right, from 3 experiments for RPE-1, n=149, 110, 124, 132, from 2 experiments for HeLa K and n=44, 76, from 2 experiments for U2OS cells).
e, f, Orthogonal method (1 μM NMS-P715, MPS1i) to generate lagging chromosomes shows a similar non-core NE assembly defect as observed with the nocodazole block-and-release protocol (Fig. 1a). e, Top: Experimental scheme. Bottom: Representative images of RPE-1 and HeLa K cells. Note that in RPE-1 cells and U2OS cells there is a near absence of non-core protein on lagging chromosomes, irrespective of the method to generate them. However, in HeLa K cells, the effect is slightly less penetrant. ~60% of lagging chromosomes lack detectable non-core protein recruitment, ~15% display strongly reduced levels (scored as labeled but shown in gray bar as “reduced”, see f) , and ~25% display a clear signal (Nup133), but often only covering part of the circumference of the lagging chromosome. These subtle differences are likely explained by differences in spindle organization between cell lines (see Fig. 3 and Extended Data Fig. 6, 7, 8 below). f, Quantitation of the results in different cell lines (n=56, 78 for RPE-1, from 2 experiments; n=75, 174 for HeLa K, from 3 experiments). Scale bars, 10 μm.
g, h, Chromatin bridges (arrows) formed after nocodazole release or after partial depletion of the condensin SMC239, display core-only NE protein assembly. g, Images of an RPE-1 cell after release from a nocodazole block (representative of 30 DNA bridges, from 5 experiments). h, Top: Experimental scheme for generating chromosome bridges by partial SMC2 depletion. Bottom: Images of an emerin-GFP expressing RPE-1 cell. Percentage of cells with the indicated staining pattern are on the lower right (n=30, from 2 experiments). Scale bars, 10 μm. Note that the DNA bridges are uniformly depleted for non-core (LBR) proteins, with no evidence for a gradient (i.e. increased LBR away from the central spindle) as might be expected for the chromosome separation checkpoint hypothesis15,40. We also note that interphase chromatin bridges were previously reported to have an altered NE protein composition including reduced levels of lamin B1 and NPCs41, which is consistent with our findings here.
