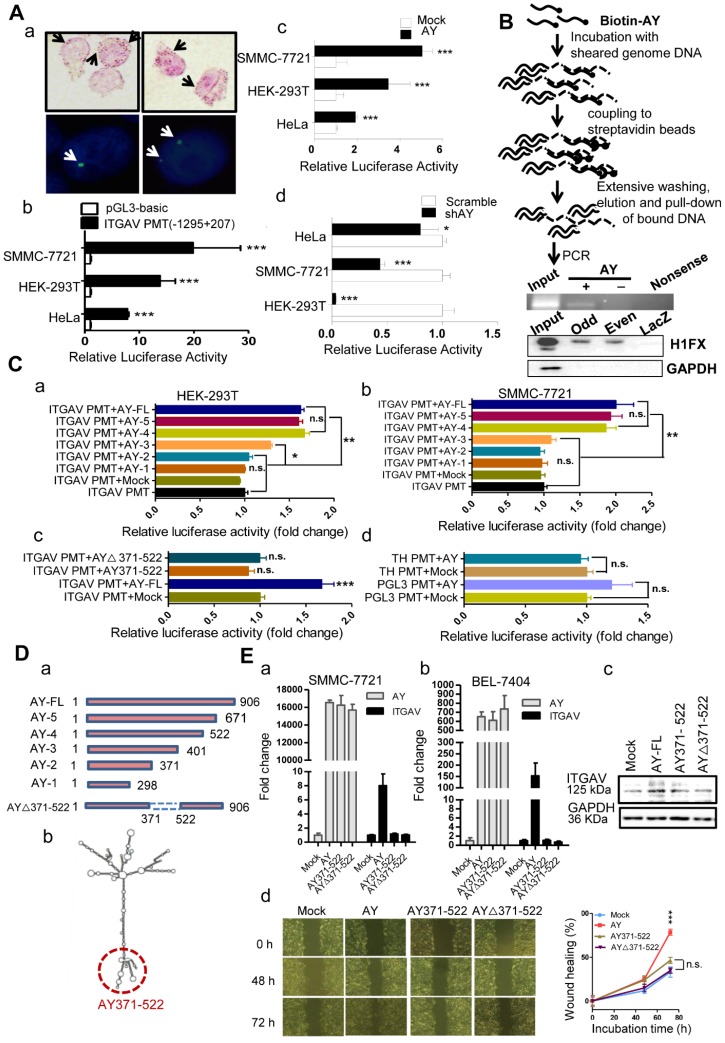
Figure 5.
AY up-regulated ITGAV transcription. A. Representative micrographs of AY in situ hybridization and fluorescent in situ hybridization (a). Arrows indicate hybridization spots. Transcription activity analysis of the ITGAV core promoter region (-1295 ~ +207) in 3 cell lines by luciferase reporter assay (b). Effect of AY expression on transcriptional activity of the ITGAV promoter using luciferase reporter assay (c & d). B. A schematic diagram of the Biotin-AY pull-down experiment (upper). PCR identification of IGTAV promoter sequence in the DNA complex pulled down by biotinylated AY. Western analysis of H1FX in the complex pulled down by biotinylated AY probes (lower). Odd, odd pool, Even, even pool of AY probes. LacZ probe as negative control. C. Effect of AY truncation on ITGAV promoter activity in HEK-293T and SMMC-7721 cells by luciferase reporter assay (a & b). Analysis of AY371-522 regulation on ITGAV promoter activity (c). The effect of AY overexpression on the unrelated pGL3 promoter and human tyrosine hydroxylase promoter (TH PMT) (d). PMT, promoter; ∆, deletion mutation. D. A schematic diagram of AY truncations that were constructed into pcDNA3.1b for overexpression (a). Predicted secondary structure for AY by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (b). The AY371-522 domain is marked by a red dotted circle. E. Quantitative comparative analysis of ITGAV expression between cells overexpressing full-length AY, AY371-522, AY△371-522, and mock control by qRT-PCR and Western blotting (a, b & c). Wound closure was analyzed in cells overexpressing full-length AY, AY371-522, and AY△371-522 (d). Data are representative of three independent repeats. *, P < 0.05; **, P < 0.01; ***, P < 0.001.