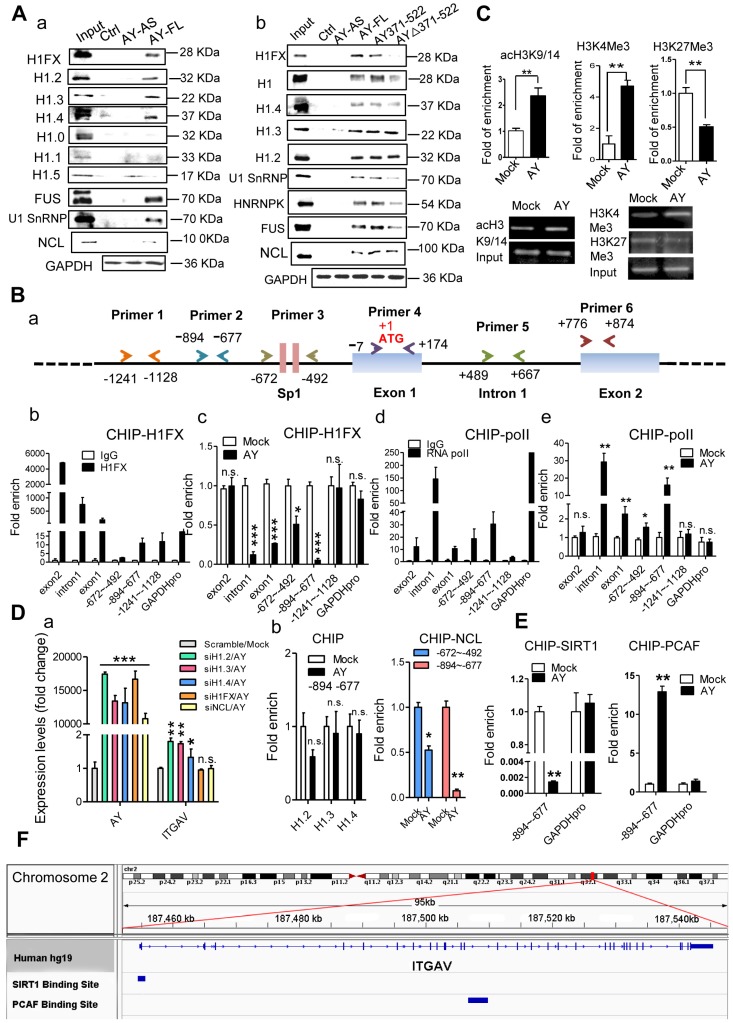
Figure 6.
AY interacted with H1FX. A. Analysis of histone 1 proteins, NCL, or U1SnRNP in AY RNA pull-down assays. B. A schematic diagram of primer design for ChIP analysis of the ITGAV promoter (a). ChIP analysis of H1FX and RNA polymerase II (RNA pol II) binding on the ITGAV promoter regions in BEL-7404 cells overexpressing AY (b-e). GAPDH promoter (GAPDH pro) and IgG served as a negative control. C. ChIP-qPCR/PCR analysis of acH3K9/14, H3K4Me3, and H3K27Me3 occupancy on the ITGAV promoter (-894 ~ -677) in BEL-7404 cells overexpressing AY. D. QPCR analysis of AY and ITGAV expression levels in cells silenced for H1 variants and overexpressing AY (a). H1.2, H1.3, H1.4, or NCL interaction with the ITGAV promoter (-894 ~ -677) was analyzed by ChIP in cells with AY overexpression (b). E. ChIP-qPCR analysis of the enrichment of PCAF and SIRT1 in BEL-7404 cells overexpressing AY. F. ChIP-Seq reads from NCBI GEO database (GSE94403, GSE15735) and alignment to human genome and transcriptome (GRCh37/hg19, Ensemblv71). Data are representative of three independent repeats. *, P < 0.05; **, P < 0.01; ***, P < 0.001.