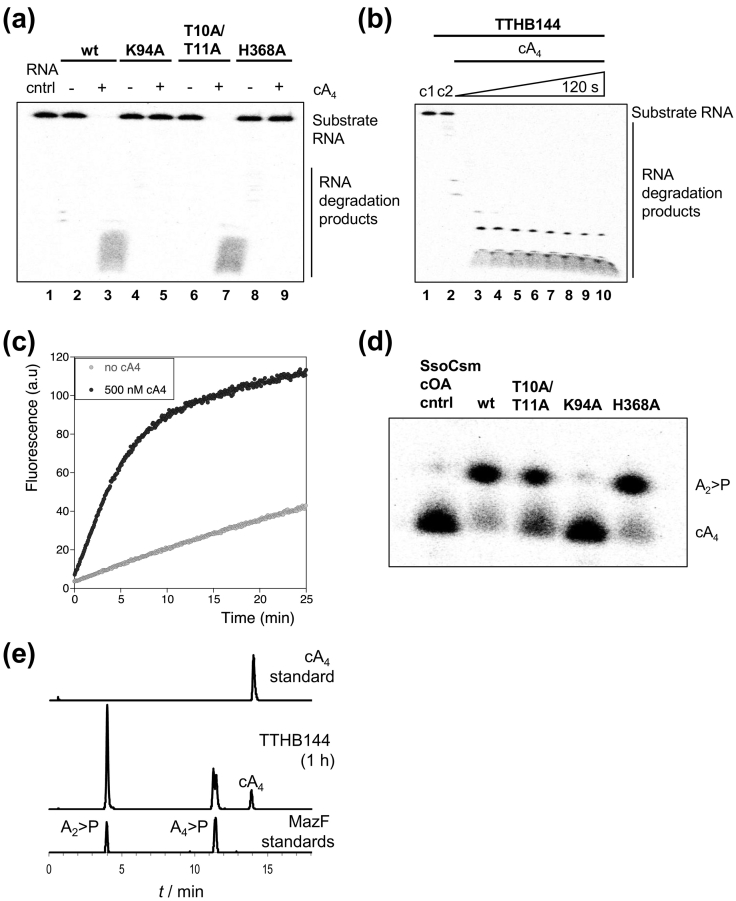
Fig. 2.
RNA degradation and cA4 cleavage occur in separate domains of TTHB144. (a) Phosphorimage of denaturing PAGE visualizing the degradation of 50 nM radiolabeled A1 RNA, as previously described [21], by TTHB144 (0.5 μM dimer), its CARF domain variants K94A and T10A/T11A and the HEPN domain variant H368A. The reaction was incubated at 70 °C for 60 min in pH 8.0 buffer containing 20 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA and 3 units SUPERase•In™ inhibitor, before quenching by phenol–chloroform extraction. RNA was cleaved by wild-type (wt) protein and the T10A/T11A variant in the presence of 1 μM cA4, but not by the K94A or H368A variants. (b) Phosphorimage of denaturing PAGE visualizing degradation of 50 nM radiolabeled RNA by TTHB144 (1 μM dimer) when incubated with 20 μM cA4 at 70 °C. Control reactions incubating RNA with buffer (c1) or RNA with protein in the absence of cA4 (c2) are shown. All of the substrate RNA was degraded within 15 s (lane 3). (c) Plot of fluorescence emitted when RNaseAlert™ substrate (1.5 μM; Integrated DNA Technologies) was degraded by 125 nM dimer TTHB144 in the absence or presence of 500 nM cA4 at 50 °C. Fluorimetry was carried out in a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) with excitation and emission wavelengths set to 490 and 520 nm, respectively. (d) Phosphorimage of denaturing PAGE visualizing degradation of 400 nM radiolabeled cA4 generated using SsoCsm complex, as previously described [21], by TTHB144 (4 μM dimer) and variants at 70 °C for 120 min. cA4 was degraded to a slower migrating product. (e) High-resolution liquid chromatography mass spectrometry of cA4 produced using the SsoCsm complex and cleavage products generated on incubation with TTHB144 (40 μM dimer) at 70 °C. cA4 (~ 16 μM; top panel) was degraded to intermediate and product species (middle panel) with identical retention times to A4 > P and A2 > P, respectively (bottom panel). A4 > P and A2 > P standards were generated using the E. coli MazF toxin as previously described [8].