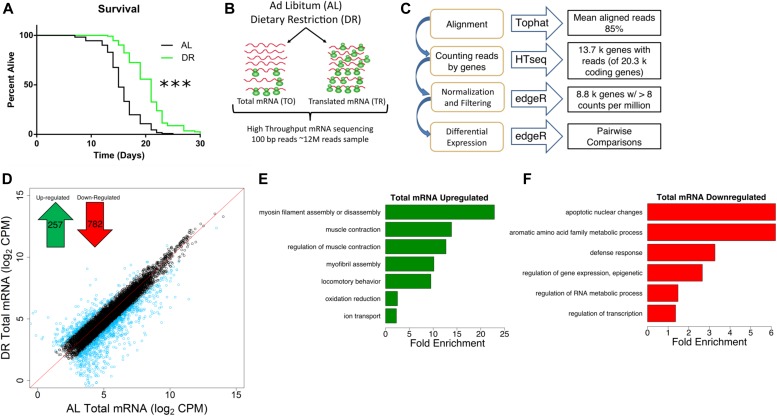
Figure 1. Transcriptional mRNA regulation under DR in C. elegans.
(A) DR increased median life span by 40% compared with well-fed conditions. P < 0.0001, n = 120, log-rank test. (B) Schematic representation of experimental setup combining mRNA-seq and polysome profiling to quantify gene expression. At day 1 of adulthood, nematodes were placed on AL or DR plates for 4 d. Total (TO) and translated (TR) mRNA were extracted from whole worm populations in biological quadruplicate and subjected to high-throughput sequencing. (C) Schematic representation of deep sequencing analysis. Reads were aligned to the C. elegans reference genome WS220 using Tophat and were counted using HTseq. Normalization, filtration, and differential expression were performed by edgeR. A total of 8,301 genes were quantified. (D) Protein-coding genes are differentially expressed under DR on the transcriptional level as determined using reads from total mRNA. Each circle represents expression for a specific gene. Black circles plotted near the red line represent genes with the same or similar expression between diets, whereas blue circles represent genes with twofold or greater change. Units are log2 values of counts aligned per million reads sequenced (CPM). Inset, 257 protein-coding genes were up-regulated and 782 genes were down-regulated significantly (FDR < 0.05) twofold or more under DR. (E, F) Biological GO terms significantly (*P < 0.05, modified Fisher's exact test) enriched among genes transcriptionally up-regulated or down-regulated under DR, respectively.