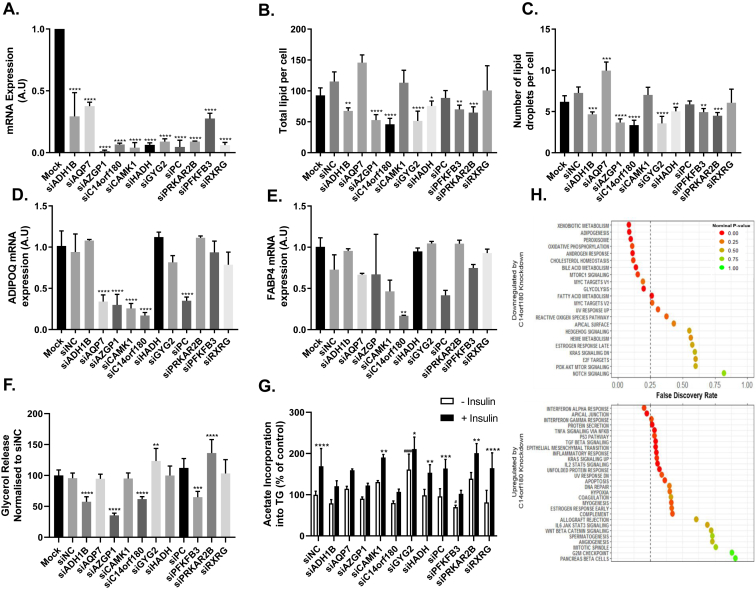
Figure 1.
Impact of candidate gene knockdown in adipose derived human mesenchymal stem cells (hMSCs) on lipids per cell, gene expression and lipid turnover. siRNAs targeting the indicated genes or siNegative Control (siNC) were transfected on Day -1 (A–E and H) or on Day 8 (F–G) of differentiation alongside a Mock transfection. For Day -1 knockdown, cells were lysed or fixed and mRNA or lipid was quantified on day 9 of differentiation. For day 8 knockdown, glycerol in the media or lipid synthesized in the cells was measured at day 13. A. mRNA quantification of the siRNA target gene compared with mock in transfected cells. B. Total lipid per cell or C. Total lipid droplets per cell were quantified after lipid staining and normalized per nucleus. n = 3 per group. D. ADIPOQ or E. FABP4 mRNA quantification after target gene knockdown as indicated on the x-axis. n = 3–7 per group. F. Basal lipolysis measured as glycerol release into the media n = 8. G. De novo lipogenesis over 3 h with or without the addition of insulin. Lipogenesis was measured via radiolabeled acetate incorporation into the cell. H. Global transcriptome profiles on siC14orf180 or siNC transfected samples were subjected to Gene Set Enrichment Analysis (GSEA). The upper panel shows gene sets downregulated and the lower panel gene sets upregulated by siC14orf180 knockdown compared to siNC. Statistical analysis carried out for B-F was target siRNA versus siNC. Statistical analysis for G carried out was (−) insulin versus (+) insulin significance denoted by * and siNC (−) insulin versus target siRNA (−) insulin significance denoted by #. One-way ANOVA with Dunnetts post hoc test was carried out for all experiments shown. *P ≤ 0.05, **P ≤ 0.01,***P ≤ 0.001, ****P ≤ 0.0001, #P ≤ 0.05, ###P ≤ 0.001.