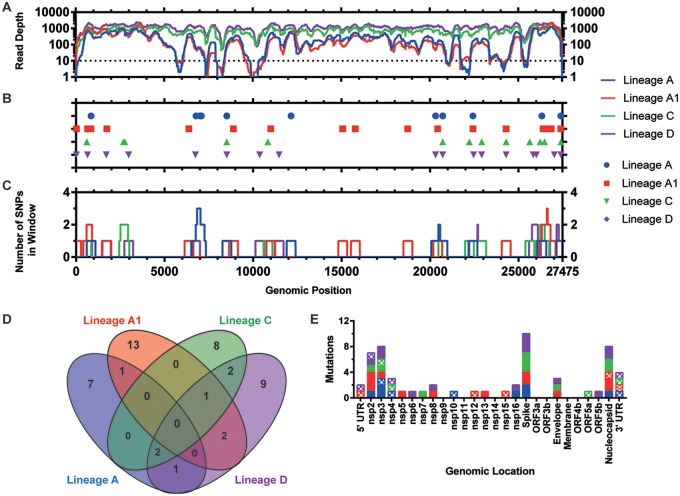
FIG 4.
Distribution of consensus level mutations across attenuated egg-passaged virus genomes. (A) Sequencing coverage of aligned final egg-passaged virus reads to the M41-CK genome. The dotted line indicates positions where consensus and minor variants have not been called. Sites with no coverage have been plotted as having a coverage of one. Totals of 529, 111, 13, and 11 sites in lineages A, A1, C, and D, respectively, had no coverage (B) Consensus-level mutations observed in the attenuated viruses relative to M41-CK EP5. Each point represents a nucleotide substitution; insertions/deletions have not been plotted. Totals of 11, 17, 13, and 17 consensus-level mutations were identified in lineages A, A1, C, and D, respectively. Consensus could not be called for 2,858, 2,959, 18, and 18 positions, respectively, due to the minimum coverage threshold (inclusive of sites with no coverage). (C) Number of consensus-level mutations occurring within a 499-nucleotide window, 249 bp on either side of the midpoint position. (D) Venn diagram of consensus-level mutations occurring at the same position in more than one virus. (E) Synonymous and nonsynonymous mutations for each virus, for each genomic location. Nonsynonymous mutations are shown as solid colors, and synonymous mutations are shown in a checkered pattern. Here the 5ʹ UTR is defined as the genomic sequence upstream of the start of nsp2, while the 3ʹ UTR is defined the genomic sequence downstream of the nucleocapsid gene. Mutations occurring in these untranslated regions have been plotted as synonymous mutations.