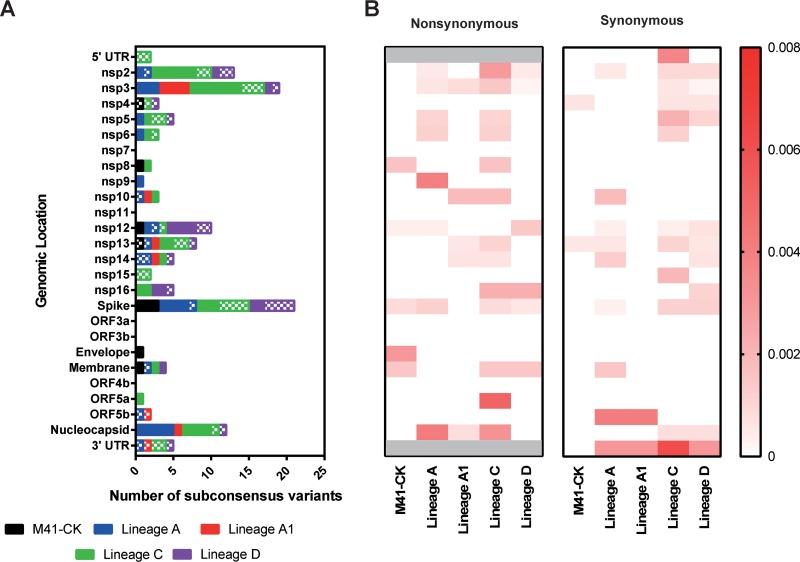
FIG 7.
Numbers of nonsynonymous and synonymous subconsensus variants per genomic location and their rate of occurrence. Variants were called in each of the five viruses against the M41-CK consensus sequence. (A) Number of NS and S subconsensus mutations per genomic location. NS mutations are shown in solids, while S mutations are shown in a checkered pattern. Mutations occurring in the 5ʹ and 3ʹ UTR have been plotted as S mutations. (B) Count of NS/S variants was divided by the length of that genomic location to calculate the rate of variation per genomic location relative to length (base pairs). Variants occurring within the 5ʹ and 3ʹ UTR have been plotted as S mutations and have been shaded out on the NS heat map.