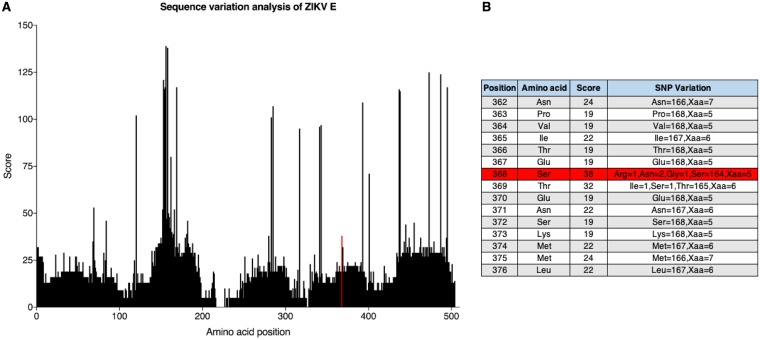
FIG 5.
Sequence alignment of ZIKV E protein. (A) A sequence alignment and a consensus sequence were generated from 173 publicly available sequences from the Virus Pathogen Resource. Each amino acid position was then assigned a polymorphism score based on Crooks et al. (46). The polymorphism score represents the normalized entropy of an observed allele distribution. Amino acid scores can have values of 0 (no observed polymorphism) to 439 (20 alleles and an indel, ∼4.7% frequency each). The red line indicates amino acid position 368. (B) The residue at site 368 (highlighted in red) has a polymorphism score of 38, where the majority of amino acids (n = 164) are serines. Of note, Xaa indicates a missing or ambiguous amino acid.