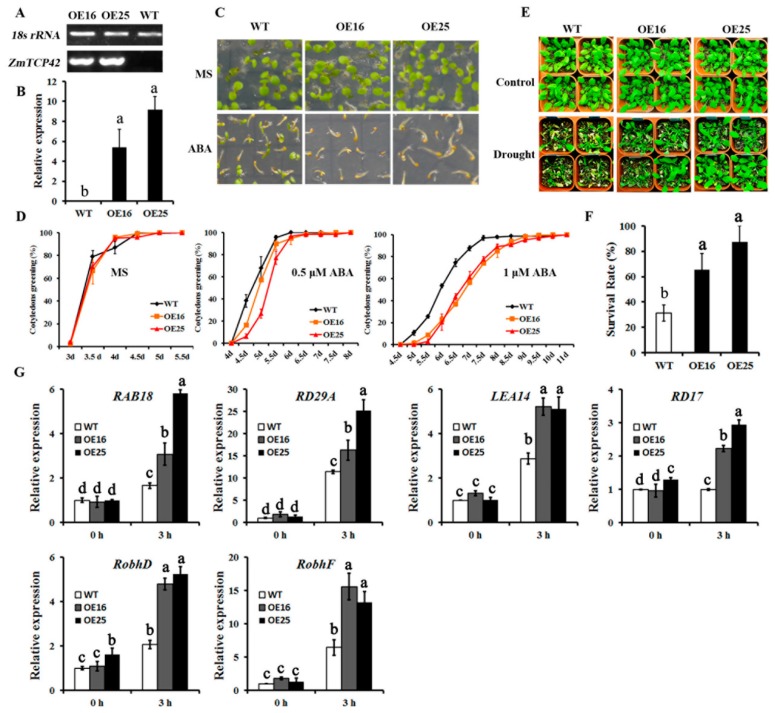
Figure 6.
Analysis of wild-type (WT) and ZmTCP42-overexpressing transgenic Arabidopsis plants. (A) RT-PCR analysis of ZmTCP42 transcript levels in ZmTCP42 transgenic lines. (B) RT-qPCR analysis of ZmTCP42 transcript levels in ZmTCP42 transgenic lines. (C) Seed germination of the ZmTCP42 transgenic lines and in the wild-type, in response to abscisic acid (ABA). Germination rates with 1 μM ABA for 5 days, defined by cotyledon greening, in wild-type ZmTCP42-OE16 and ZmTCP42-OE25, compared to the MS medium alone. (D) Statistical analysis of a green seedling’s rate of ZmTCP42 transgenic lines and wild-type grown on Murashige and Skoog (MS) medium with 0, 0.5 μM, and 1 μM ABA. (E) Drought tolerance of the ZmTCP42 transgenic Arabidopsis plants. Photographs were taken before and after the drought treatment, followed by 3 days of rewatering. (F) Statistical analysis of survival rates after the drought-stress treatment. The average survival rates and standard errors were calculated from three independent experiments. (G) RNA levels of stress-responsive genes in the ZmTCP42 transgenic lines and in the wild-type, in response to drought stress. Total RNA was obtained from 3 weeks-old seedlings treated by 3 h of dehydration stress and was analyzed by RT-qPCR, using the gene-specific primers listed in Table S2. The mean value and standard error were calculated from three replicates after normalization to ACTIN2. The RNA levels in the wild-type grown under non-stress conditions was taken as 1.0. Significant differences were calculated by one-way ANOVA with Duncan’s multiple test (SAS Institute, Inc., Cary, NC, USA). Different letters indicate a significant statistical difference between the sample means at p = 0.05, while means with the same letters were not significantly different.