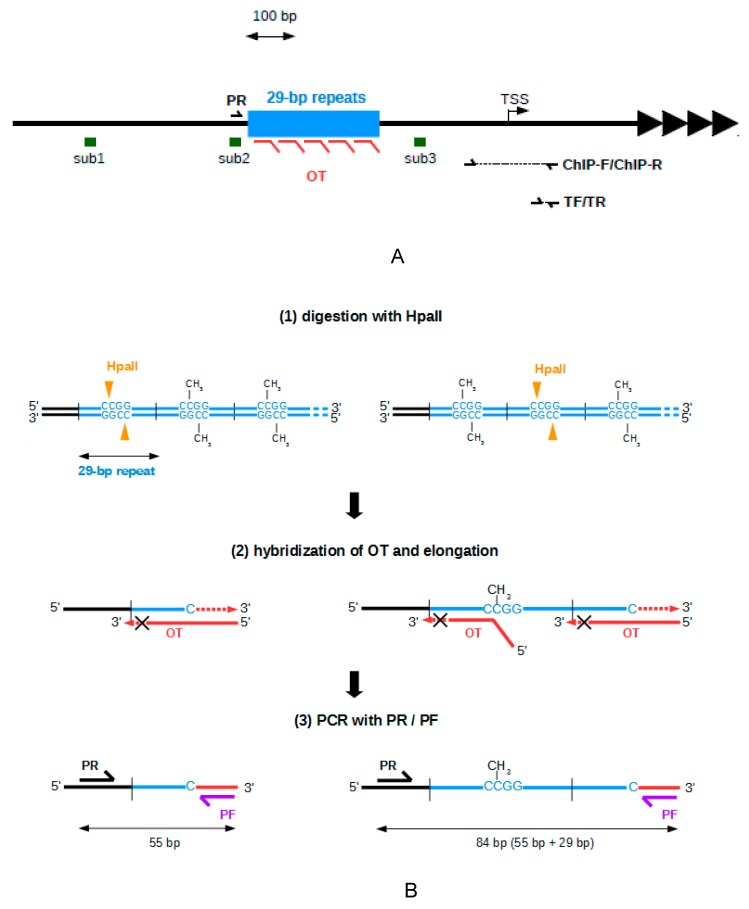
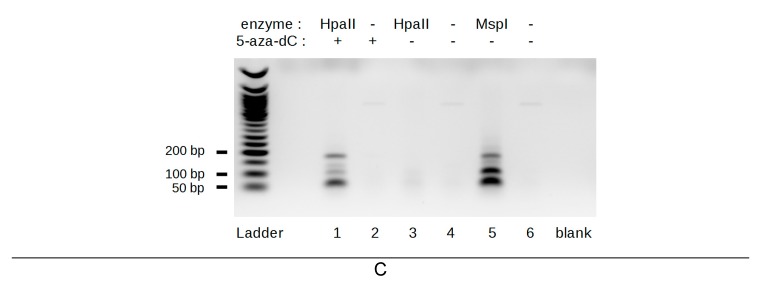
Figure 1.
The 29 bp repeats of the subtelomeric CpG islands are hypermethylated in HeLa cells. (A) Schematic representation of the subtelomeric region containing CpG island. Arrowheads indicate telomeric repeats, and blue box represents the subtelomeric 29 bp repeats. The subtelomeric telomere repeat containing RNA (TERRA) transcription start site (TSS) is indicated by a black arrow. The locations of the targets for RNA guides (sub1, sub2, and sub3) are indicated by green bars. The binding sites of the Reverse Primer (PR), Oligonucleotide Template (OT) (red), and the primer pairs used for the chromatin immunoprecipitation (ChIP) (ChIP-Foward/ChIP-Reverse (ChiP-F, ChiP-R)) and primers for the qPCR of TERRA, Foward (TF) and Reverse (TR) are indicated. Sequences amplified by these two latter primer pairs are indicated by dashed lines. (B) Schematic representation of the application of the method of analysis of the cytosine methylation at 29 bp repeats. In this example, two DNA molecules with two different patterns of cytosine methylation in the 29 bp repeats are represented (blue lines). Other 29 bp repeats are represented by dotted blue lines. (1) Genomic DNA is digested by the methylation-sensitive endonuclease HpaII (the HpaII recognition sites located within each 29 bp repeat are indicated by orange arrowheads). (2) After hybridization of the 3′ region of the oligonucleotide OT (red line) to the sequence adjacent to the HpaII recognition sites of the 29 bp repeats, DNA polymerase-catalyzed elongation (represented by dotted red lines) is performed in which the 5′ region of OT serves as template. OT is blocked at its 3′ end by dideoxycytidines to prevent its own elongation represented by crossed dotted red lines. (3) PCR is carried out with the Reverse Primer PR (black, complementary to a region upstream of the 29 bp repeats) and Forward Primer (PF) (purple, complementary to sequence added at the 3′ end of the cleaved DNA molecules). In this example, PCR products of two different sizes are obtained depending of the methylation pattern of the region. (C) Validation of the method was performed by analysis of 1 µg of genomic DNA extracted from HeLa cells untreated or treated with 5-aza-dC was undigested or digested by HpaII (methyl sensitive) or MspI (methyl insensitive). A sample without DNA (blank) was used as negative control. Elongation products (1/20 of the reaction) were amplified by PCR (35 PCR cycles). PCR products were run on a 1.5% BET-agarose gel. Ladder is the 50 bp DNA ladder (New England BioLabs).