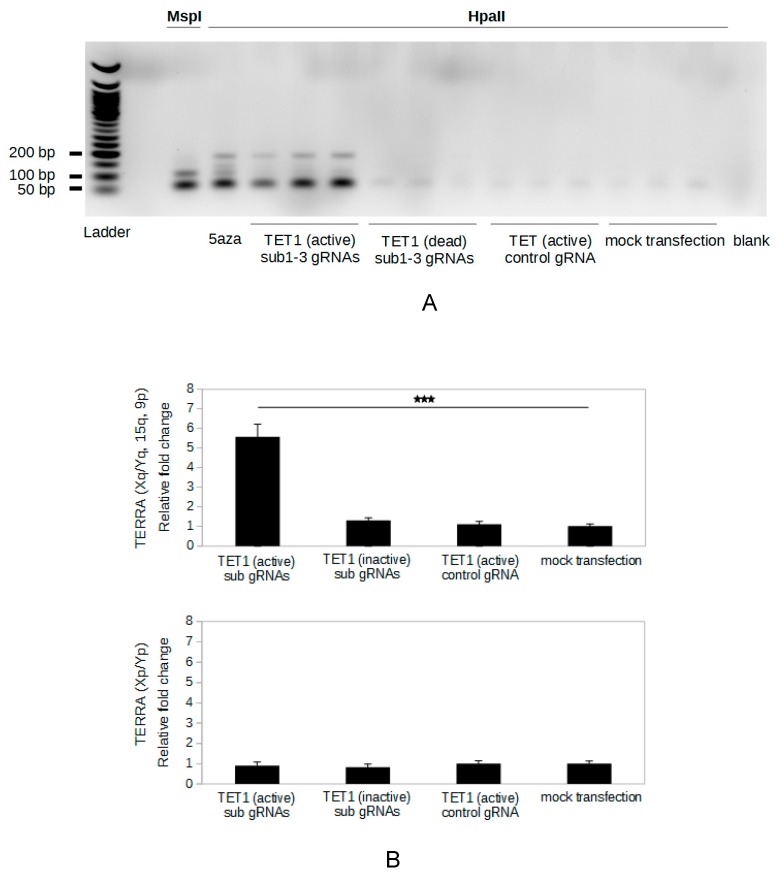
Figure 2.
Demethylation of the subtelomeric 29 bp repeats by the CRISPR-dCas9-TET1 system is associated with an up-regulation of TERRA. HeLa cells were transfected with the CRISPR-dCas9-TET1 system with TET1 active or catalytically dead and subtelomeric gRNAs (sub1-3) or control gRNA. Genomic DNA and RNA were extracted 96 h post-transfection. Mock transfected HeLa cells were used as control. (A) The methylation of the 29 bp repeats was analyzed using the method described in Figure 1B: 1 µg of genomic DNA was digested with HpaII, 1/20 of the elongation reaction was amplified by PCR (35 PCR cycles), and PCR products were run on a 1.5% bromure ethidium (BET)-agarose gel. Three independent transfection experiments were done for each condition. Genomic DNA from HeLa cells treated with 5-aza-dC and digested by HpaII (5aza) or untreated and digested with MspI were used as positive controls. A sample without DNA (blank) was used as negative control. Ladder is the 50 bp DNA ladder (New England BioLabs). (B) TERRA from subtelomeres Xq, Yq, 15q, 9p and TERRA from subtelomeres Xp and Yp were quantified by RT-qPCR (reverse transcription-quantitative PCR) using TF/TR and Xp-R/Xp-F primer pairs, respectively. Levels were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, and all values were compared to mock transfected sample. The bars are average values from three biological and two technical replicates for each sample. Error bars are standard deviations. p values were calculated by paired two-tailed Student’s t-test (n = 3). *** p < 0.001.