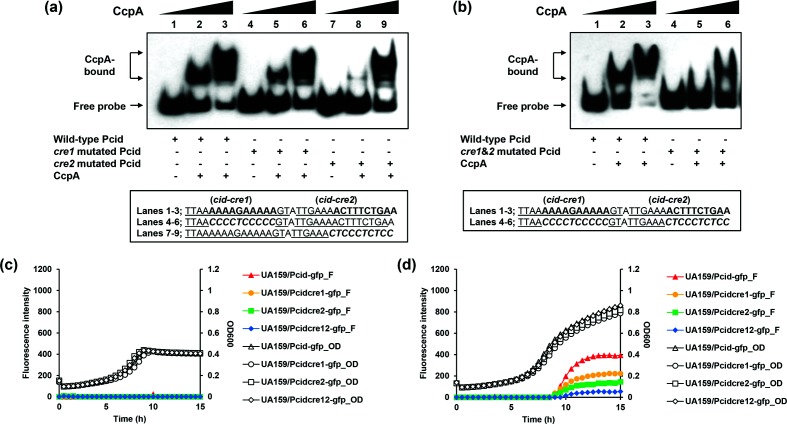
Fig. 4.
Effects of putative cre site mutations in the cid promoter region. (a–b); EMSA. Underlined letters indicate two putative CcpA-binding (cid-cre1 and cid-cre2) sequences. Italic letters indicate the mutated sequence regions. EMSA was performed with 1 fmol biotinylated cid promoter (Pcid) regions with mutation in the putative cre sequences and various amounts of purified His-CcpA (0, 0.78125 or 1.5625 pmol). The reactions were run on a non-denaturing polyacrylamide gel and the signal observed by chemiluminescence. (a) Lanes 1–3 contain biotinylated Pcid wild-type DNA probes; lanes 4–6 contain biotinylated Pcid-cre1 mutated DNA probes; and lanes 7–9 contain biotinylated Pcid-cre2 mutated DNA probes. (b) Lanes 1–3 contain biotinylated Pcid wild-type DNA probes; lanes 4–6 contain biotinylated Pcid-cre1&2 mutated DNA probes. (c–d); Effects of cre mutations on cid expression over growth. The S. mutans UA159 (wild-type) strains contained pDL278 carrying the gfp gene driven by wild-type and mutated Pcid. The strains were grown in a chemically defined medium (FMC) supplemented by either a low [11 mM, (c)] or high level [45 mM, (d)] of glucose. Relative gfp fluorescence intensity (coloured lines; F) and OD600 (black lines; OD) were monitored on a plate reader (see Methods for details). The results are the average values for at least three biological replicates performed in triplicate.