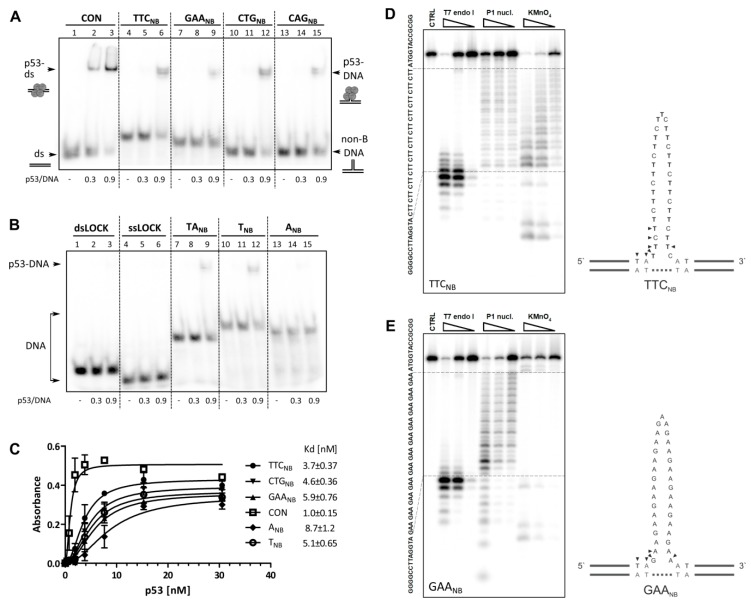
Figure 1.
Full length p53 binding to non-B DNA formed by repetitive sequences. Analysis of p53 binding to DNA molecules containing various sequences and structures. DNA sequences are provided in Table S1. In panels (A,B), radiolabeled oligonucleotides (1 pmol) were bound by p53 in binding buffer in the presence of non-specific DNA competitor (75 ng, BSK/SmaI) and analyzed on a 6% 0.5× TBE polyacrylamide gel. The p53/DNA ratios are given per p53 tetramer. (A) DNA molecules are slipped strand mimicking substrates formed by trinucleotide repeat sequences (11 repeats of TTC, GAA, CTG or CAG) and a duplex p53 consensus sequence (CON). (B) DNA molecules formed by TANB, TNB, ANB, and DNA sequence nonspecific for p53 (dsLOCK, ssLOCK). (C) p53 binding to biotinylated oligonucleotides by ELISA, portrayed are mean values and SD. The dissociation constants (Kd) calculated per p53 tetramer are indicated. (D,E) Polyacrylamide gels showing resolved products of TTCNB (D) and GAANB (E) enzymatic or chemical cleavage with increasing concentration of T7 endonuclease I (left), P1 nuclease (middle) and potassium permanganate followed by piperidine treatment (right). Right panels show schematic drawings of formed structures. Small arrows indicate preferential cleavage by T7 endonuclease I.