Figure 1. Establishing conditions of azidohomoalanine (AHA) labelling to examine protein synthesis in mice following intraperitoneal administration.
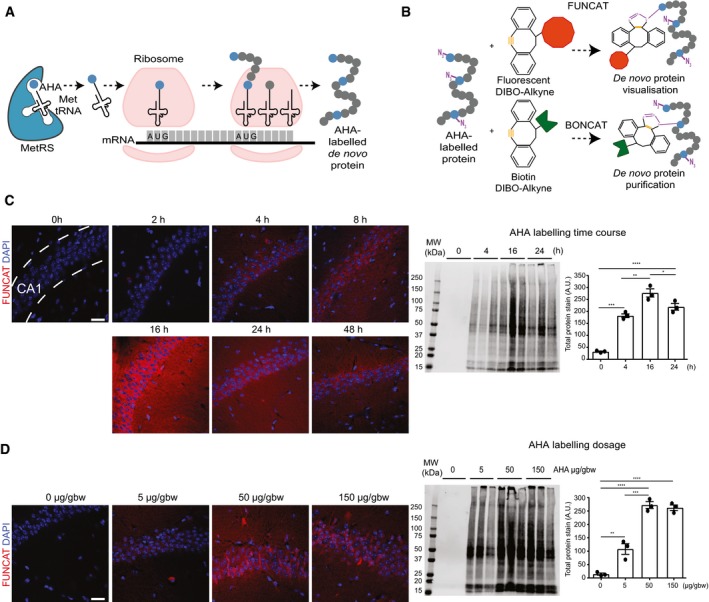
- Schematic representation of AHA labelling as a tool to identify newly synthesised proteins. De novo synthesised proteins are labelled with AHA at the amino‐terminal and internal methionine residues using the endogenous translational machinery.
- AHA‐labelled proteins can be covalently bonded through reaction of the azide group (purple) of AHA with the alkyne group (orange) of tags and either visualised using fluorescent non‐canonical amino acid tagging (FUNCAT) or purified using bio‐orthogonal non‐canonical amino acid tagging (BONCAT) for further analysis.
- FUNCAT visualisation in wild‐type (WT) mice treated for varying time periods with 50 μg AHA per gram body weight (gbw). AHA incorporation can be observed as early as 4‐h post‐injection in the CA1 region of the hippocampus and is still observed 48‐h post‐injection. Western blot analysis of AHA‐labelled proteins purified from whole hemisphere (without the cerebellum) with BONCAT reveals that maximal AHA labelling occurs approximately 16‐h post‐injection (n = 3 mice, one‐way ANOVA, Tukey's multiple comparison test).
- AHA‐labelled proteins in WT mice treated with ascending concentrations of AHA were visualised using FUNCAT. Scale bar: 40 μm. Western blot of AHA‐labelled proteins purified from whole hemisphere (without the cerebellum) using BONCAT reveals that maximal AHA labelling occurs when mice are administered 50 μg AHA/gbw (n = 3 mice, one‐way ANOVA, Tukey's multiple comparison test).
