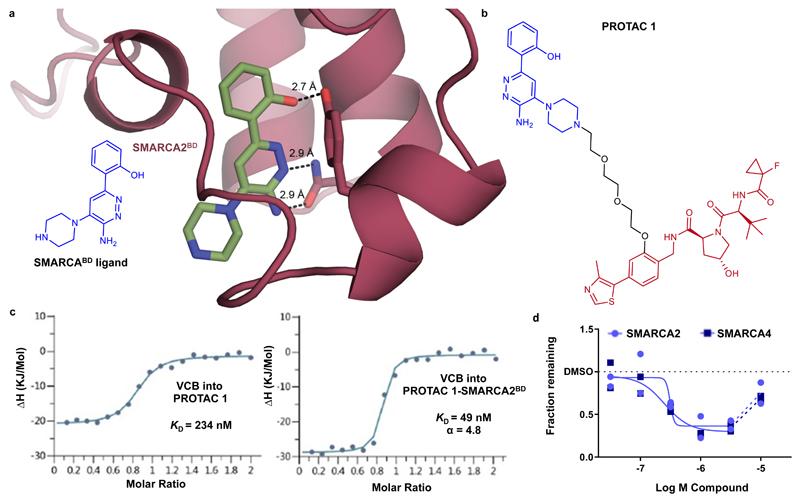
Figure 1. Rational design and evaluation of a partial degrader of SMARCA2 and SMARCA4, PROTAC 1.
a) 2D chemical structure of a SMARCABD ligand and crystal structure of this ligand in complex with SMARCA2BD. The piperazine ring was selected as an exit vector for PROTAC linkage as it is directed into solvent away from the binding site. b) 2D chemical structure of PROTAC 1 c) Inverse ITC titrations of VCB into PROTAC 1 (left) and VCB into the preformed PROTAC 1–SMARCA2BD complex (right), n = 2. PROTAC 1 binds VCB with higher affinity when in complex with SMARCA2BD and is therefore cooperative, α = 4.8. d) Degradation of SMARCA2 and SMARCA4 in MV-4-11 cells following treatment with PROTAC 1, analysed via capillary electrophoresis (see online methods). For SMARCA2 and SMARCA4, maximal degradation is ~65 % and ~70%, and DC50 is 300 nM and 250 nM, respectively. Data represent means from two biologically independent experiments, ± S.E.M.