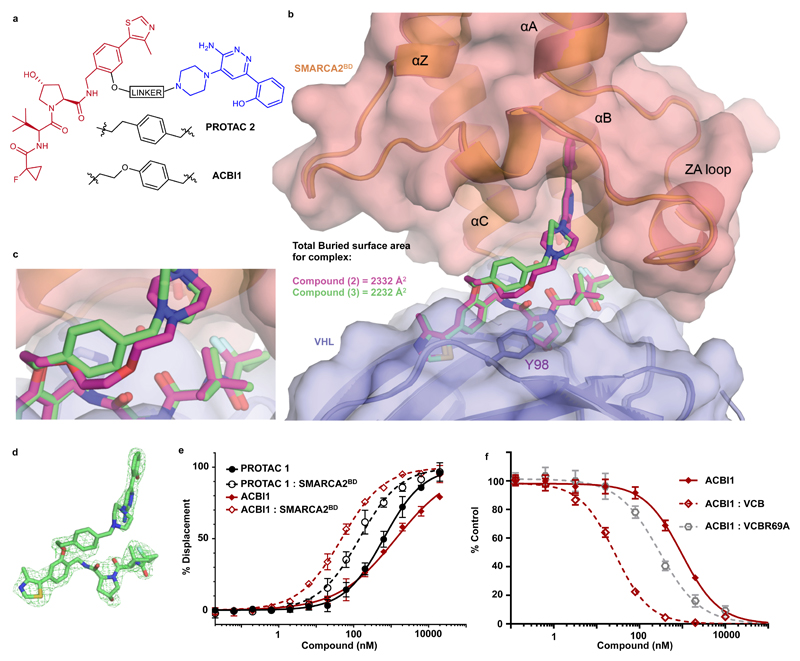
Figure 3. Ternary co-crystal structure of SMARCA2BD: PROTAC 2: VCB and biophysical data validate a rational design strategy.
a) 2D chemical structures of PROTAC 2 and ACBI1 b) Overlays of ternary crystal structures of VCB: PROTAC 1 : SMARCA2BD (orange, PROTAC carbons in magenta) and VCB: PROTAC 2 : SMARCA2BD (2.35 Å, salmon, PROTAC carbons in green). Near identical ternary complexes are formed by both PROTACs; with the phenyl ring of PROTAC 2 in close proximity to Y98 of VHL. c) Overlays show that the constrained 1,4-disubstituted phenyl ring of PROTAC 2 (green carbons) accurately recapitulates the linker geometry observed previously for PROTAC 1 (magenta carbons). d) Fo-Fc omit map (green meshes) of PROTAC 2 prior to ligand modelling contoured at 3.0 σ with a carve radius of 2.2 Å. e) Fitted curves from Fluorescence Polarization competition assays measuring displacement of a VHL peptide by PROTACs in the presence or absence of SMARCA2BD. ACBI1 forms more cooperative and stable complexes compared with PROTAC 1. Curves are a best fit of means from n = 3 biologically independent experiments, ± S.E.M. f) Fitted curves for TR-FRET assays measuring displacement of a biotinylated SMARCA2BD probe by PROTAC alone, in complex with VCB or in complex with an R69A variant of VCB. A significant rightward shift when using VCBR69A vs VCB highlights the importance of this residue in ternary complex formation in solution. Curves are a best fit of means from n = 3 biologically independent experiments, ± S.D.