Figure EV4. DNA duplex processing by Sgs1 together with RPA and Top3‐Rmi1 at high‐salt conditions.
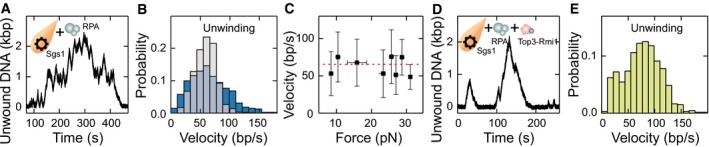
- dsDNA processing events observed for Sgs1 in the presence of RPA at high‐salt conditions.
- Histograms of unwinding velocity of Sgs1 in the presence of RPA at high‐salt conditions. Shown are the velocities in the presence of 20 nM RPA (gray) and 200 nM RPA (dark blue). Mean unwinding velocities are v = 59 ± 2 bp/s (N = 860) and v = 65 ± 3 bp/s (N = 1,095) for 20 and 200 nM RPA, respectively.
- Force dependence of the Sgs1 unwinding (black squares) (N = 1,095) velocities in the presence of 200 nM RPA. Red dashed lines represent mean values of the velocities, and error bars represent the standard deviation of the mean.
- dsDNA processing events observed for Sgs1 in the presence of RPA and Top3‐Rmi1 at high‐salt conditions.
- Histogram of the unwinding velocity of Sgs1 in the presence of RPA and Top3‐Rmi1. The mean unwinding velocity was v = 77 ± 4 bp/s (N = 678).
