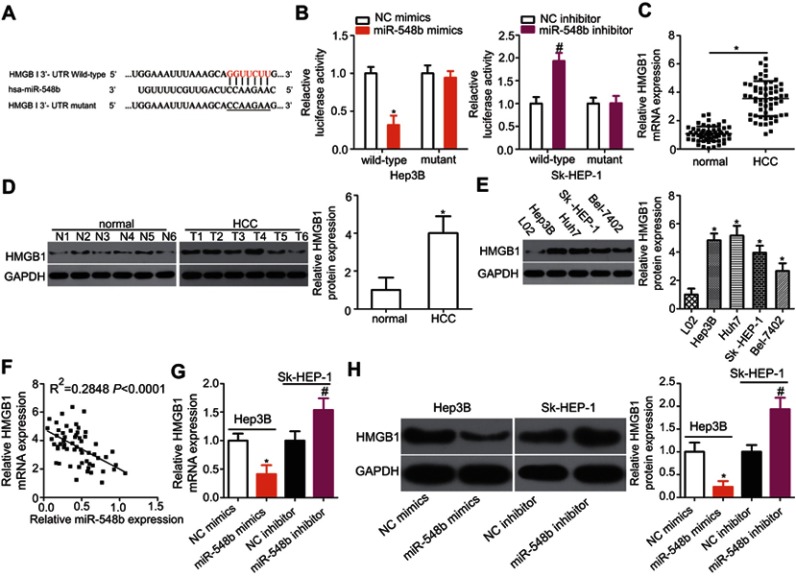
Figure 4.
HMGB1 mRNA is a direct target of miR-548b in hepatocellular carcinoma (HCC) cells. (A) The putative binding site of miR-548b in the 3′-UTR of HMGB1 mRNA. The mutant site is also presented. (B) Forced miR-548b expression reduced the luciferase activity yielded by the plasmid carrying the wild-type 3′-UTR in Hep3B cells. Downregulation of miR-548b in SK-HEP-1 cells increased the luciferase activity generated by the plasmid harboring the wild-type binding site. *P<0.05 as compared with NC mimics. #P<0.05 as compared with the negative control (NC) inhibitor. (C) The expression of HMGB1 mRNA was evaluated in 59 pairs of HCC and normal paracarcinoma tissue samples by reverse-transcription quantitative (RT-q) PCR. *P<0.05 as compared with normal paracarcinoma tissue samples. (D) Western blot analysis was conducted to measure HMGB1 protein expression in several pairs of HCC and normal paracarcinoma tissue samples. *P<0.05 as compared with normal paracarcinoma tissues. (E) Western blotting was applied to determine the HMGB1 protein expression in four HCC cell lines – Hep3B, Huh7, SK-HEP-1, and Bel-7402 – with an immortalized normal human liver epithelial cell line (L02) as a control. *P<0.05 as compared with L02 cells. (F) A negative correlation between HMGB1 mRNA and miR-548b levels among HCC tissue samples was confirmed by Spearman’s correlation analysis. R2=0.2848, P<0.001. (G) HMGB1 mRNA was quantified by RT-qPCR in Hep3B and SK-HEP-1 cells after transfection with miR-548b mimics or the miR-548b inhibitor, respectively. *P<0.05 as compared with NC mimics. #P<0.05 as compared with the NC inhibitor. (H) The protein expression of HMGB1 in Hep3B cells was obviously decreased by transfection with miR-548b mimics, whereas the miR-548b inhibitor significantly increased HMGB1 protein expression in SK-HEP-1 cells. *P<0.05 as compared with NC mimics. #P<0.05 as compared with the NC inhibitor.