Abstract
We developed a screening assay in which luciferized ID8 expressing OVA was cocultured with transgenic CD8+ T cells specifically recognizing the model antigen in a H-2b–restricted manner. The assay was screened with a small molecule library to identify compounds that inhibit or enhance T cell–mediated killing of tumor cells. Erlotinib, an EGFR inhibitor, was the top compound that enhanced T-cell killing of tumor cells. Subsequent experiments with erlotinib and additional EGFR inhibitors validated the screen results. EGFR inhibitors increased both basal and IFNγ-induced MHC class-I presentation, which enhanced recognition and lysis of tumor cell targets by CD8+ cytotoxic T lymphocytes. The ID8 cell line was also transduced to constitutively express Cas9, and a pooled CRISPR screen, utilizing the same target tumor cell/T-cell assay, identified single-guide (sg)RNAs targeting EGFR that sensitized tumor cells to T cell–mediated killing. Combination of PD-1 blockade with EGFR inhibition showed significant synergistic efficacy in a syngeneic model, further validating EGFR inhibitors as immunomodulatory agents that enhance checkpoint blockade. This assay can be screened in high-throughput with small molecule libraries and genome-wide CRISPR/Cas9 libraries to identify both compounds and target genes, respectively, that enhance or inhibit T-cell recognition and killing of tumor cells. Retrospective analyses of squamous-cell head and neck cancer (SCCHN) patients treated with the combination of afatinib and pembrolizumab demonstrated a rate of clinical activity exceeding that of each single agent. Prospective clinical trials evaluating the combination of an EGFR inhibitor and PD-1 blockade should be conducted.
Keywords: EGFR, immunotherapy, CRISPR, cytotoxic T cell, checkpoint blockade
Introduction
With the FDA approval of immune checkpoint blocking antibodies, initially targeting CTLA-4 in melanoma (1) and then for PD-1/PD-L1 in melanoma (2), NSCLC (3), head and neck cancer (4), and others (5–7), the field of medical oncology has experienced a paradigm shift in treatment modalities. Combination CTLA-4 and PD-1/PD-L1 blocking antibodies have exhibited synergistic efficacy (2,8), and numerous trials and pre-clinical development pipelines are ongoing that utilize antibodies that block one or both of these immune checkpoints in combination with additional checkpoint-blocking antibodies (LAG-3, TIM-3, TIGIT, B7-H3) or agonistic monoclonal antibodies (4–1BB, OX-40, GITR, CD40, ICOS). However, despite all these approaches, not all patients benefit from immunotherapy and, as such, additional therapeutic strategies to enhance the effects of immunotherapy are needed.
There is increasing interest in combination therapies that leverage existing technologies to increase the immunogenicity of solid tumors and augment immunotherapeutics, such as anti-PD-1/PD-L1, that are increasing being viewed as foundational reagents in the medical oncology field. Radiotherapy (9), chemotherapy (10,11), and targeted agents, such as inhibitors of HDACs (NCT02619253, NCT02437136), BRAF (NCT02818023), and VEGF (NCT00790010) have been or are currently being tested clinically in combination with immune checkpoint blockade and have been shown to increase response rates.
The repurposing of existing conventional therapeutics for combination with checkpoint blockade is an attractive strategy, given the pre-existing pharmacodynamic/pharmacokinetic and toxicology properties of such compounds. It was our goal to develop an assay that could be utilized to screen compound libraries in high-throughput for identification of immunomodulatory features. We engineered a target tumor cell line to express firefly luciferase and a model antigen. We proceeded to coculture these target cells with transgenic CD8+ T cells recognizing the model antigen, such that modulation of antigen-specific, T cell–mediated killing could be assessed by luminescence readout and would identify candidate compounds with immunomodulatory properties. The screen identified the epidermal growth factor receptor (EGFR) as an unappreciated immune-oncology target whose inhibition enhanced anti–PD-1 immunotherapy.
Materials and Methods
Cell lines.
ID8 were obtained from the laboratory of Gordon Freeman (DFCI) in 2014, MC38 were purchased from ATCC in 2015, 293T were purchased from Invitrogen in 2011, and the KrasG12D;p53–/– (KP) cell line was derived in-house from the mouse model (16) in 2016. All cell lines were confirmed to be mycoplasma negative by Charles River Research Animal Diagnostic Services using standard Quantitative Fluorescence PCR (QF-PCR) protocol. Cell lines were authenticated by short-tandem repeat (STR) profiling. Only cell lines of <20 passages were used for experiments.
Generation of the luciferized ID8 cell lines.
A firefly luciferase-OVA fusion cassette was cloned from the Lenti-LucOS vector as previously described (12) using two-step PCR (13) with primers as follows: attL1 forward: 5’-AGGCTCCTGCAGGACCATGGAAGACGCCAAAAAC-3’; attL2 reverse 5’-GAAAGCTGGGTCTCGAGCTAGCGGCCGCTTACAAG-3’; attL1-T1 forward: 5’-CCCCGATGAGCAATGCTTTTTTATAATGCCAACTTTGTACAAAAAAGCAGGCTCCTGCAGGACCATG-3’; attL2-T1 reverse: 5’-GGGGGATAAGCAATGCTTTCTTATAATGCCAACTTTGTACAAGAAAGCTGGGTCTCGAGCTA-3’. PCR products containing the lucOS open reading frame (ORF) was then inserted into the pLVX-IRES-Neo lentiviral vector (Clontech, Mountain View, CA) using Gateway® LR Clonase® II (Thermo Fisher, Waltham, MA). A renilla luciferase vector was constructed using the same protocol and also inserted into the pLVX-IRES-Neo lentiviral vector. Plasmids were transformed into One Shot® OmniMAX™ 2 competent cells according to the manufacturer’s protocol (Thermo Fisher, Waltham, MA). Clones were miniprepped (Qiagen, Valencia, CA), genotyped by PCR, sequence-verified, and transiently transfected into 293T cells to assess firefly luciferase expression. Positive clones were co-transfected into 293T cells along with d8.9 and VSV-G packaging plasmids (Addgene, Cambridge, MA). ID8-Cas9 cells were transduced with pLVX-lucOS-IRES-Neo or pLVX-rluc-IRES-Neo vectors and placed under G418 selection for seven days. Viral production and ID8 spin-fection were conducted according to the Broad Institute’s lentiviral production guidelines (14). Clonal cell lines of “lucOS” and “rluc” cell lines were generated by limiting dilution, expanded for 3–4 weeks under puromycin and G418 selection to obtain sufficient cell numbers, and verified for luciferase and OVA expression. KrasG12D;p53–/– (KP) cell lines were transduced to express lusOS or rluc constructs utilizing the same protocols.
Harvesting and activation of OT-I T cells.
C57BL/6-Tg(TcraTcrb)1100Mjb/J OT-I mice (stock #003831; Jackson labs, Bar Harbor, ME) were bred in-house. 8–12 week-old mice were sacrificed, and spleens were harvested by mechanical separation through a 40 μM filter. Red blood cells were lysed using 1X RBC lysis buffer (Biolegend, San Diego, CA). Splenic single-cell suspensions were resuspended in TruStain fcX™ (anti-mouse CD16/32; Biolegend, San Diego, CA) FcR block diluted 1:100 in FACS buffer (PBS plus 2% FBS) and incubated on ice for 15 minutes. CD8+ OT-I T cells were stained with mouse CD8 (Ly-2) microBeads for 20 minutes, washed with FACS buffer, and isolated using magnetic separation and LS columns according to manufacturer’s protocol (kit# 130–049-401; Miltenyi Biotec, San Diego, CA). CD8+ T cells were eluted into RPMI (Life Technologies, Carlsbad, CA) containing 10% FBS (HyClone, Logan, UT), penicillin (100 units/mL), and streptomycin (100 μg/mL; Life Technologies, Carlsbad, CA). OT-I CD8+ T cells were then activated with Dynabeads Mouse T-Activator CD3/CD28 beads (Life Technologies, Carlsbad, CA) for 24 hours before addition to lucOS/rluc cocultures. OT-I CD8+ T-cell purity of >95% was confirmed by flow cytometry.
OT-I CTL assay.
10,000 ID8-lucOS and 10,000 ID8-rluc cells were plated in 100 μL of cell culture media (DMEM plus 10% FBS, penicillin (100 units/mL), and streptomycin (100 μg/mL) in solid white, flat-bottomed, tissue culture-treated 96-well plates (Thermo Fisher, Waltham, MA). Overnight-stimulated OT-I CD8+ T cells were then added at the designated effector:target (E:T) ratios with or without compounds (Supplementary Table S1) in a total volume of 200 μL and concentration of 1 μM. Plates were incubated for 48 hours at 37°C and 5% CO2. After 48 hours, 100 μL of media was removed prior to the dual-luciferase assay (cat# E2940, Promega, Madison, WI). Calculation of the Fluc/Rluc ratio or of percent surviving OVA-expressing target cells ([Fluc +OT-I/Rluc +OT-I] / [avg Fluc no OT-I/avg Rluc no OT-I] x 100) was utilized where indicated for analysis of compound performance.
Briefly, 50 μL of Dual-Glo® luciferase buffer (15) was added to wells and incubated for 30 minutes at room temperature before analysis of firefly luminescence. 50 μL of Dual-Glo® Stop & Glo® buffer was then added, plates were incubated for 30 minutes, and then analyzed for renilla luminescence on an EnSpire plate reader (PerkinElmer, Waltham, MA). 203 compounds (Supplementary Table S1; details for LINCS kinase library can be found at http://lincs.hms.harvard.edu/db/sm/) were screened in high-throughput at 1 μM final concentration in duplicate in 96-well plates containing both CD8+ OT-I cytotoxic T lymphocytes (CTLs) and ID8 target cells or ID8 target cells only at E:T of 1:1. No compounds were placed in edge wells, and all plates contained multiple DMSO control cells. Performance of compounds was calculated based on ΔΔCt method using ΔΔCt = (FLucDMSO/RLucDMSO) / (FLuccompound/RLuccompound), where values >1 augment CTL killing, values ~1 have a negligible immunomodulatory effect, and values <1 inhibit CTL killing. Compounds reaching a certain threshold (top 10%) from high-throughput screen were validated with dose-response curves using the indicated drug concentrations.
OT-I IFNγ ELISA.
100 μL of the supernatants from the OT-I CTL assays described above were harvested at the 48-hour timepoint prior to the dual-luciferase assay and analyzed for IFNγ secretion with a LEGEND MAX™ Mouse IFNγ ELISA Kit (Biolegend, San Diego, CA) according to the manufacturer’s protocol. Indicated compounds were tested at 100, 50, 10, 5, and 1 nM, with DMSO only control wells with either no OT-I CTLs or E:T ratios of 1:1 and 2:1. Conditions were assayed with four replicate wells per experiment.
Flow cytometry for MHC class-I expression.
100,000 ID8-lucOS, ID8-lucOS sgEGFR KO, KrasG12D;p53–/–, and MC38 cell lines were cultured in 12-well plates in 2 mL of culture media (DMEM containing 10% FBS, penicillin (100 units/mL), and streptomycin (100μg/mL) alone or supplemented with recombinant mouse IFNγ (4 ng/mL; Biolegend, San Diego, CA) for 48 hours. Where indicated, cells were treated with erlotinib, gefitinib, or afatinib at a concentration of 100 nM for the full 48-hour time course. EGFR KO cells had loss of EGFR confirmed by Western blot prior to the assay. Cells were trypsinized, washed, and resuspended in FACS buffer (PBS plus 2% FBS) with H-2Kb-APC (clone AF6–88.5; Biolegend) at a dilution of 1:100 for 15 minutes on ice, then washed twice prior to analysis on a BD LSRFortessa with FACSDiva software (BD Biosciences, San Jose, CA). Data were analyzed using FlowJo (Ashland, OR) software version 10.0.9.
sgEGFR constructs.
The top four scoring sgRNA targeting EGFR (sequences listed in Supplementary Table S2) were ordered as oligos from IDT (Coralville, Iowa) and ligated into BsmBI site in pXPR-sgRNA-GFP-Blast expression vector (Addgene, Cambridge, MA) using Quick Ligation Kit according to manufacturer’s protocol (cat# M2200S New England Biolabs, Ipswich, MA). Plasmids were transformed into One Shot Stbl3 Chemically Competent E. coli according to the manufacturer’s protocol (cat# C737303 Thermo Fisher, Waltham, MA). Clones were miniprepped (Qiagen, Valencia, CA), genotyped by PCR, and sequence-verified. Positive clones were co-transfected into 293T cells along with d8.9 and VSV-G packaging plasmids (Addgene, Cambridge, MA). ID8-Cas9 cells were transduced with pXPR-sgEGFR-GFP-Blast and placed under G418 selection for seven days. Viral production and ID8 spin-fection were conducted according to the Broad Institute’s lentiviral production guidelines (14).
CRISPR/Cas9 screen.
ID8-lucOS cells stably expressing Cas9 were transduced with a ~8000 guide pooled sgRNA library (Supplementary Table S2) with 10 sgRNA/gene covering: 87 control genes (essential genes, oncogenes, tumor suppressor genes), 86 immune modulators (immune checkpoints, differentially regulated immune genes), 524 epigenetic regulators, 34 MHC genes, and 500 non-targeting sgRNAs. sgRNAs were expressed from the pXPR-sgRNA-2A-GFP vector (Addgene, Cambridge, MA) at MOI of 0.3 and selected for blasticidin resistance at a representation of 500 cells/sgRNA, which was maintained throughout the screen. CD8+ OT-I T-cells were harvested and pre-stimulated as in the plated-based compound screen and added to T175 flasks with monolayers of sgRNA-transduced ID8-lucOS cells at an E:T ratio of 1:1. Control flasks with no OT-I T cells added were also passaged in parallel. Cell cultures were maintained for 72 hours, at which point, live and dead ID8-lucOS cells were harvested for isolation of genomic DNA.
Genomic (g)DNA from cell pellets was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Carlsbad, CA) and was concentrated using Genomic DNA Clean & Concentrator (Zymo Research, Irvine, CA), both according to manufacturers’ protocol. Twelve μg gDNA (250X representation for 8000 sgRNAs at 6 ρg DNA/cell) was amplified using Titanium Taq DNA Polymerase (Clontech, Mountain View, CA) in a one-step PCR reaction with the following parameters: 95°C 1 minute, [95°C 30 seconds, 64°C 30 seconds, 72°C 30 seconds] X 22 cycles, 72°C 5-minute first step with F2/R2 primers. PCR products were verified on a DNA1000 Bioanalyzer (Agilent, Santa Clara, CA) and ~350 bp bands were gel-purified using the QIAquick Gel Extraction Kit (Qiagen, Carlsbad, CA). PCR products were diluted to 10 ng/μL, pooled, and sequenced on a NextSeq machine (Illumina, San Diego, CA).
In vivo validation.
C57BL/6J mice (stock #000664; Jackson labs, Bar Harbor, ME) were challenged subcutaneously with 500,000 MC38 colon cancer cells on their flanks and enrolled on-study when tumors reached 50 mm3. Mice were treated with vehicle plus IgG2a isotype control (10 mg/kg; Bio X Cell, West Lebanon, NH), anti–PD-1 (10 mg/kg; clone RMP1–14; Bio X Cell, West Lebanon, NH), afatinib (10 mg/kg; Selleck, Houston, TX), combination anti–PD-1 (10 mg/kg) and afatinib (10 mg/kg), or combination anti–PD-1 (10 mg/kg), afatinib (10 mg/kg), and anti-CD8α (200 μg; clone 53–6.7; Bio X Cell, West Lebanon, NH). Animals received intraperitoneal (IP) injections of anti–PD-1 on days 5, 8, and 12 and afatinib on days 6, 7, 8, 9, and 10 (as indicated). Depleting anti-CD8α was administered two days prior to first anti–PD-1 treatment. Mice used in experiments were 7–8 weeks of age at time of tumor challenge. Endpoint was considered to be when tumors reached a size of 2000 mm3 or as mandated by institutional guidelines due to development of necrotic lesions. Mice were monitored every 2–3 days and tumors measured with digital calipers. All animal studies were conducted in accordance with, as well as with the approval of, the Institutional Animal Care and Use Committee of Dana-Farber/Harvard Cancer Center.
Afatinib and pembrolizumab combination therapy.
Retrospective medical record and image review of patients with recurrent and/or metastatic squamous-cell carcinoma of the oral cavity, oropharynx, hypopharynx, or larynx (SCCHN) who received combination afatinib and pembrolizumab at National Taiwan University Hospital between November 1, 2016 and September 30, 2017 with follow-up through March 30, 2018 were evaluated. Exclusion criteria included prior treatment with afatinib, pembrolizumab, or nivolumab as a monotherapy or prior treatment with other anti-cancer agents in combination with afatinib or pembrolizumab. Disease status was assessed by MRI or CT scan, and responses were reviewed by a radiologist according to RECIST 1.1 criteria. In all, 41 patients were eligible for analysis, with clinical annotation and treatment regimen available in Supplementary Table S3. Patient studies were conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS). Studies were performed after approval by the Institutional Review Board of the National Taiwan University Hospital (NTUH) (Taipei, Taiwan). Investigators obtained informed written consent from all subjects.
Data analysis.
The following denote statistical significance: *p-value <0.05; **p-value <0.01; ***p-value <0.001. Flank tumor growth curves were analyzed using two-way ANOVA, all bar graphs were analyzed using unpaired Student’s t-test, and survival experiments used the log-rank Mantel-Cox test for survival analysis. Statistics were calculated using PRISM 7.01 (GraphPad, La Jolla, CA).
Results
Development of OT-I Immune Oncology (IO) assay
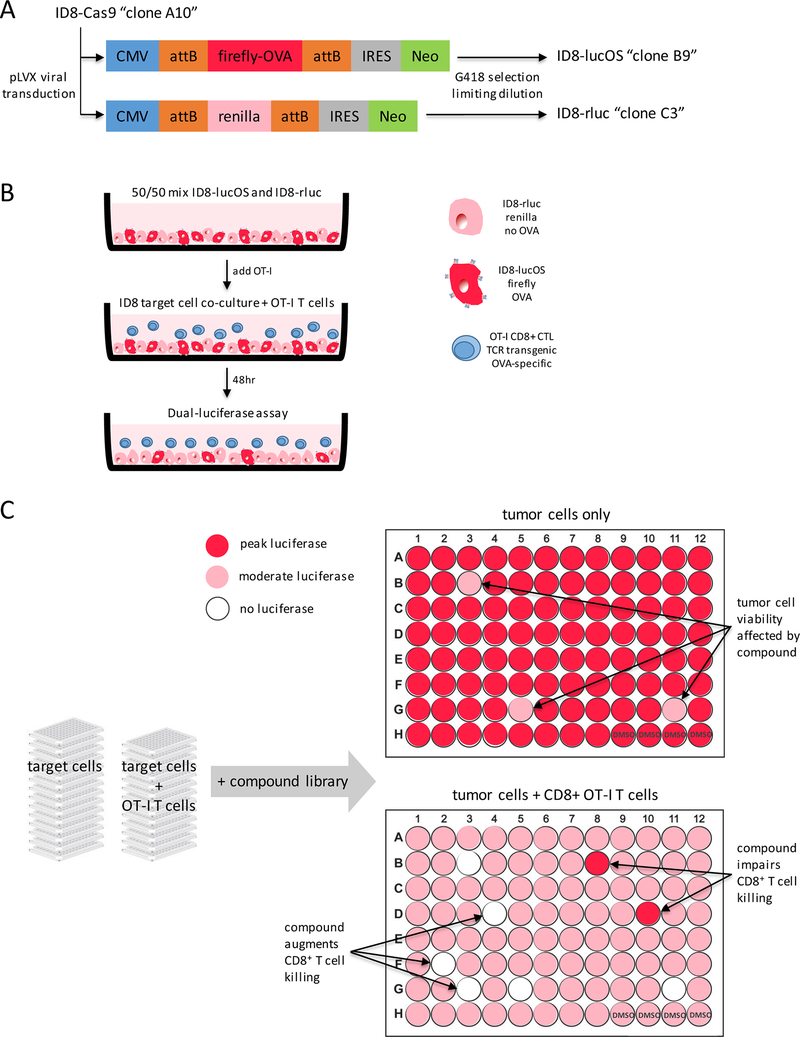
We selected the ID8 murine serous ovarian carcinoma cell line due to its constitutive expression of MHC class-I, MHC haplotype compatibility with C57BL/6J mice, and IFNγ-induced upregulation of PD-L1 (Supplementary Fig. S1). ID8 cells were transduced with pLVX vectors to express either firefly luciferase and OVA model antigen (“lucOS”) or renilla luciferase and no model antigen (“rluc”) (Fig. 1A). Target ID8 cell lines were mixed at a 1:1 ratio and cocultured with CD8+ T cells isolated from the spleens of OT-I TCR-transgenic mice. OT-I mice express transgenic TCRα-V2 and TCRβ-V5 genes such that all CD8+ T-cell receptors recognize chicken ovalbumin residues 257–264 (SIINFEKL) in the context of H-2Kb (17). Target cell–T cell cultures were incubated with compounds for 48hrs. and then analyzed by dual-luciferase assays, where changes in firefly signal relative to controls would indicate modulation of T-cell killing by compound treatment (Fig. 1B and C).
Figure 1: OT-I CTL screen design.
(A) ID8-Cas9 serous ovarian carcinoma cell line was transduced with a pLVX vector expressing either firefly luciferase fused to a model antigen peptide or renilla luciferase and no antigen. Clonal cell lines were generated using G418 selection for Neo cassette expression and limiting dilution. (B) 10,000 ID8-lucOS and 10,000 ID8-rluc were co-plated into wells of 96-well tissue culture plates. OT-I TCR transgenic CD8+ T cells were then plated on top of ID8 cells. Total volume/well was 200 μL and was incubated for 48hrs. at 37°C and 5% CO2 prior to analysis by dual-luciferase assay. (C) OT-I assay was screened in high-throughput to evaluate compounds for immunomodulatory effects on antigen-specific tumor cell killing by cytotoxic T lymphocytes (CTLs). Inclusion of ID8-lucOS and ID8-rluc provided in-plate normalization controls to determine non-specific growth inhibition or induction of apoptosis by screen compounds.
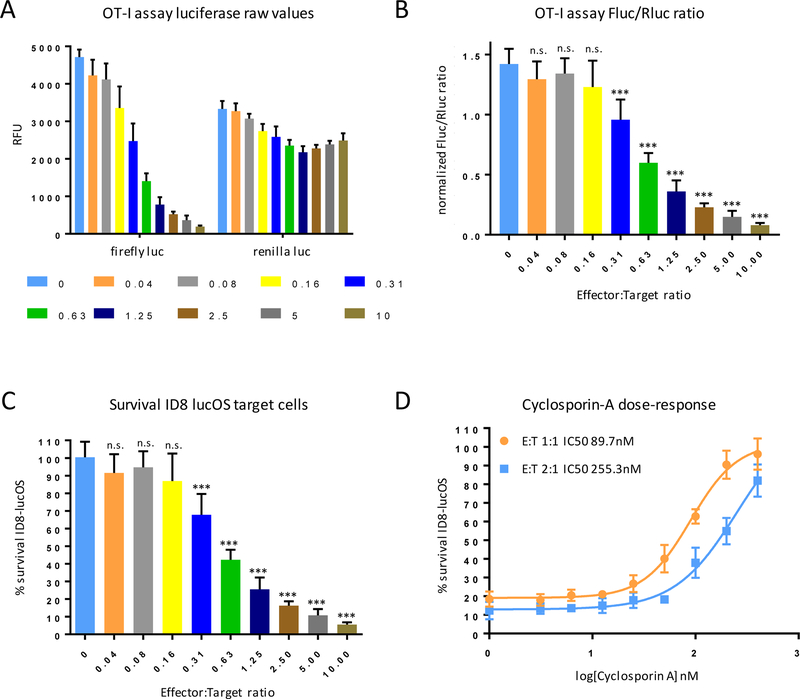
The renilla luciferase signal remained relatively constant across wells, regardless of the number of OT-I T cells added to coculture. However, a dramatic loss of firefly luciferase signal with increasing Effector:Target ratios, indicating that OT-I CD8+ T cells selectively kill lucOS ID8 cells in an antigen-dependent manner, while sparing rluc ID8 cells (Fig. 2A). Simple calculation of the Fluc/Rluc ratio or of percent surviving OVA-expressing target revealed that an Effector:Target ratio of ~0.5 was sufficient to observe ~50% killing (Fig. 2B and C). The OT-I IO assay was validated with cyclosporin-A, a well-established inhibitor of CD8+ T-cell effector function (18). As expected, cyclosporin-A inhibited T cell–mediated killing of antigen-expressing target cells in a dose-dependent manner, consistent with published IC50 values (Fig. 2D).
Figure 2: OT-I assay validation.
(A) 10,000 ID8-lucOS and 10,000 ID8-rluc were plated in 96-well plates with OT-I CD8+ T cells at the indicated E:T ratio to assess antigen-specific tumor cell killing. (B) Analysis of normalized luciferase ratios and (C) calculation of % survival of target ID8-lucOS cells at the indicated E:T ratios. (D) Dose-response curves for calcineurin inhibitor cyclosporin-A used as a control compound to validate assay performance. Experiments were performed at least twice with six replicate wells per condition. Data for bar graphs calculated using unpaired Student’s t-test with *p<0.05, **p<0.01, and ***p<0.001 and presented as mean with SD.
OT-I IO assay pilot screen and hit validation
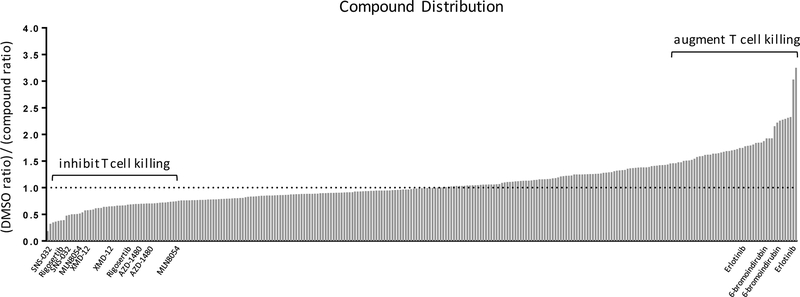
We first screened our OT-I IO assay with a focused library of kinase inhibitors from the Harvard Medical School NIH LINCS Center (Supplementary Table S1). Compounds were screened at a 1 μM concentration, a dose at which nearly a third of the compounds caused non-specific loss of viability in both antigen-expressing lucOS and control rluc ID8 cells. These compounds were removed from further analysis (Supplementary Fig. S2). Screen results were analyzed based on the normalized firefly/renilla luciferase ratio in DMSO control wells relative to compound-treated wells (Fig. 3). Compounds were considered hits if they fell in the top or bottom 10% of compounds and scored in all replicate plates. Compounds inhibiting OT-I T-cell killing had ratios <1, inert compounds had ratios ~1, and compounds that augmented T-cell killing displayed ratios >1. The CDK9 inhibitor SNS-032, PLK1 inhibitor Rigosertib, aurora kinase A inhibitor MLN8054, JAK2 inhibitor AZD-1480, and aurora kinase inhibitor XMD-12–1 (19,20) all inhibited T cell–mediated target cell lysis (Fig. 3). The GSK-3β inhibitor 6-bromoindirubin and EGFR inhibitor erlotinib were the only two compounds that significantly augmented T-cell killing.
Figure 3: Compound distribution from the OT-I screen.
Normalized firefly/renilla luciferase ratios relative to DMSO-only control wells. Plates were screened in duplicate, and compounds were considered “hits” only if they scored in replicate plates. Compounds inhibiting OT-I T-cell killing had ratios <1 (JAK2 inhibitor, CDK9 inhibitor, PLK1 inhibitor), inert compounds had ratios ~1, and compounds that augmented T-cell killing display ratios >1 (EGFR inhibitor, GSK-3β inhibitor).
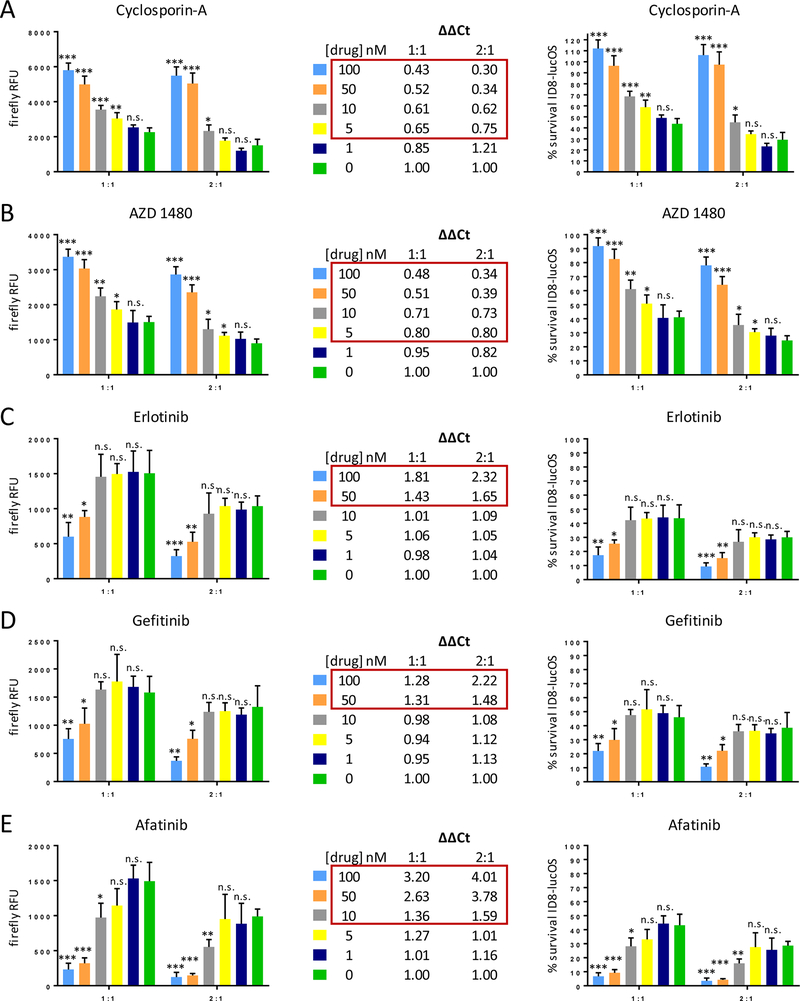
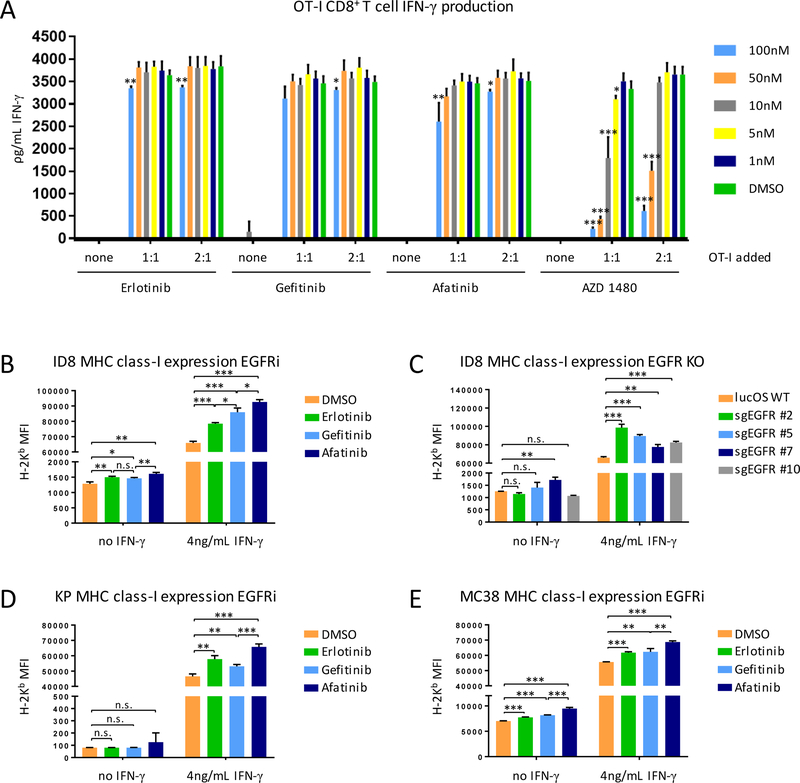
Hits from the initial larger compound screen were validated in the OT-I IO assay with dose-response curves. The JAK2 inhibitor AZD-1480 significantly inhibited T-cell killing with similar kinetics to cyclosporin-A, down to low nM concentrations (Fig. 4A and B). However, because of synergistic application with immune checkpoint blockade, compounds that augmented T-cell killing were of interest. Upon further testing, the GSK-3β inhibitor 6-bromoindirubin and other GSK-3β–specific and GSK-3 inhibitors only modestly augmented T-cell killing (Supplementary Fig. S3). The EGFR inhibitor erlotinib, however, was confirmed to augment T cell–mediated tumor cell lysis (Fig. 4C). To determine if this effect was erlotinib-specific or EGFR-specific, we also tested gefitinib, an alternative EGFR-specific ATP competitive inhibitor, and afatinib, an irreversible EGFR inhibitor of different chemotype. All three EGFR inhibitors significantly augmented OT-I T-cell killing and, in the case of afatinib, resulted in lysis of almost all OVA-expressing ID8 target cells even at concentrations down to 10 nM (Fig. 4C-E). The mutant selective EGFR tyrosine kinase inhibitor (TKI) osimertinib exhibited enhanced killing that was inferior to erlotinib, gefitinib, and afatinib (Supplementary Fig. S3). Osimertinib has activity against wild-type EGFR only at high concentrations (21).
Figure 4: Validation of the compound screen results.
(A) Cyclosporin-A control dose-response exhibited predicted inhibition of OT-I T-cell killing. (B) AZD 1480 (JAK2 inhibitor) performed similarly to Cyclosporin-A. (C-E) Erlotinib (EGFR inhibitor) and two other EGFR inhibitors (gefitinib, afatinib) were assayed across a dose range and confirmed screen result that inhibition of EGFR augments antigen-specific T-cell killing. Data presented as raw firefly luciferase (OVA-expressing ID8) values for two different E:T ratios (left), relative firefly luciferase values that account for drug impacts on ID8 survival irrespective of T cell-mediated killing (middle), and % survival of ID8-lucOS target cells (right). Experiments were conducted at least twice with similar results and in replicates of four wells per condition. Data for bar graphs calculated using unpaired Student’s t-test with *p<0.05, **p<0.01, and ***p<0.001 and presented as mean with SD.
To eliminate the possibility that the EGFR sensitivity observed in our assay was an artifact of the ID8 cell line, we also performed a CTL assay in a cell line derived from the KrasG12D/p53–/– C57BL/6J lung adenocarcinoma model (Supplementary Fig. S4) (16). This KP cell line was transduced with the same vectors to stably express Cas9 and the lucOS construct and cocultured with OT-I CD8+ T cells. OT-I T cell–mediated lysis of OVA-expressing KP cells was significantly enhanced by EGFR inhibitors erlotinib, gefinitib, and afatinib and inhibited by cyclosporin-A, further confirming our initial ID8 screen result (Supplementary Fig. S4).
Tumor cell-intrinsic effect of EGFR inhibition
Cell culture media from the OT-I IO assay was harvested from wells following 48hrs. of culture, with a range of EGFR inhibitors and the JAK2 inhibitor AZD-1480, and assessed for IFNγ secretion by ELISA, which was used as a proxy for OT-I T-cell effector function. As expected, escalating doses of AZD-1480 significantly inhibited IFNγ secretion in a dose-dependent manner (Fig. 5A). None of the EGFR inhibitors affected IFNγ secretion, leading us to conclude that the immunomodulatory effect of EGFR inhibition in our assay was not due to T cell-intrinsic effects.
Figure 5: EGFR inhibition enhances T-cell killing via a tumor cell-intrinsic mechanism.
(A) ELISA of IFNγ secretion by OT-I CD8+ T cells cocultured for 48hrs. with 10,000 cells at the indicated E:T ratios with the indicated compound treatment. (B-E) Expression of MHC class-I by the indicated target tumor cells after inhibition of EGFR with three different compounds of varying chemotypes or sgRNA targeting EGFR. Experiments were conducted at least twice with similar results and in replicates of four wells per condition. Data for bar graphs calculated using unpaired Student’s t-test with *p<0.05, **p<0.01, and ***p<0.001 and presented as mean with SD.
It has been previously reported that EGFR inhibitors increase basal and IFNγ-induced expression of MHC class-I expression in human keratinocytes (22), leading us to investigate if this mechanism might explain the increased T cell–mediated killing following treatment with EGFR inhibitors in our assay. We observed that erlotinib, gefitinib, and afatinib all significantly increased both basal expression of MHC class-I by ID8 tumor cells and MHC class-I expression induced by physiological IFNγ (Fig. 5B). GFR inhibitor-induced upregulation of MHC class-I expression also correlated with performance of the varying EGFR inhibitors in the OT-I IO assay. The irreversible inhibitor afatinib was superior to ATP competitive inhibitors erlotinib and gefitinib. The same cell line transduced with multiple different sgRNA targeting EGFR (Supplementary Fig. S5) also exhibited increased basal and IFNγ-induced expression of MHC class-I (Fig. 5C). KrasG12D;p53–/– lung adenocarcinoma (Fig. 5D) and MC38 colon cancer (Fig. 5E) cell lines responded to EGFR inhibitor treatment by significantly increasing surface MHC class-I.
A CRISPR/Cas9 screen independently identifies EGFR as immunomodulatory
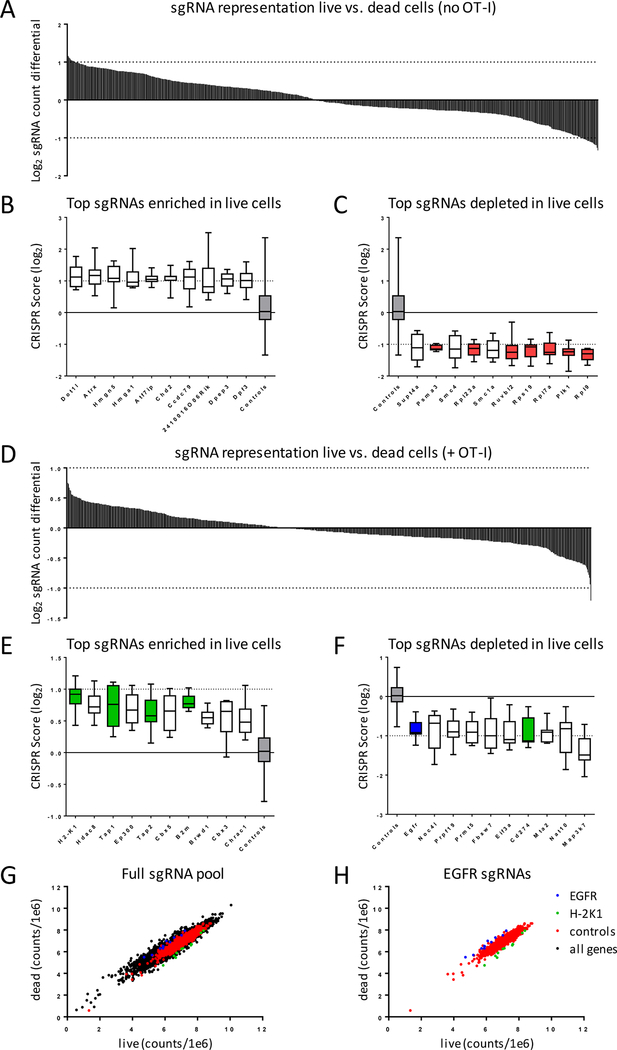
Our OVA-expressing ID8 target cell line was also engineered to constitutively expresses the Cas9 gene, enabling us to transduce these cells with a sgRNA library and perform the OT-I IO assay in a pooled format. We utilized a library of ~8,000 sgRNAs comprised of 87 control genes (essential genes, oncogenes, tumor suppressor genes), 86 immune modulators (immune checkpoints, differentially regulated immune genes), 524 epigenetic regulators, and 34 MHC genes at a coverage of 10 sgRNA per gene and also included 500 non-targeting sgRNAs (Supplementary Table S2). ID8 lucOS cells were transduced with the lentiviral library and cultured at a representation of 500 cells/sgRNA for 72hrs. in the presence or absence of OT-I effector CD8+ T cells. In the absence of OT-I T cells, as expected, sgRNAs targeting essential genes were preferentially depleted in surviving cells (Fig. 6A-C, red bars). With the addition of OT-I T cells (Fig. 6D), we expected that positive control sgRNAs that targeted immunosuppressive mechanisms, such as PD-L1, would enhance CTL killing, whereas negative control sgRNAs targeting MHC class-I processing and presentation genes should inhibit CTL killing. SgRNAs targeting H2-K1, Tap1, Tap2, and B2m scored as four of our top seven genes enriched in live cells following coculture with OT-I CTLs (Fig. 6E, green bars). SgRNAs targeting our positive control targeting PD-L1 were preferentially depleted in live cells, indicating that loss of this immunosuppressive surface receptor sensitized the ID8 cells to T cell–mediated killing (Fig. 6F, green bar). SgRNAs targeting EGFR were also preferentially depleted from surviving ID8 cells, indicating that loss of EGFR sensitized tumor cells to T cell-mediate killing. EGFR scored as #10 out of 731 genes depleted in live cells (Fig. 6F-H). Top ranking sgRNAs targeting EGFR were used to make individual stable EGFR KO cell lines, which were also sensitized to OT-I T cell–mediated killing across a wide range of Effector:Target ratios, validating our pooled CRISPR screen results (Supplementary Fig. S5)
Figure 6: CRISPR/Cas9 screen identifies sgRNAs targeting EGFR as sensitizing tumor cells to T-cell killing.
(A-C) ID8-lucOS cells alone or (D-H) cocultured at E:T of 1:1 with OT-I T cells were incubated for 72hrs. After which, genomic DNA was isolated, and sgRNA sequences were deconvoluted by NGS. (E,G) sgRNAs targeting genes enriched and (F,H) sgRNAs targeting genes depleted in OT-I cultures. CRISPR score was defined as the average log2 fold-change in abundance of sgRNAs for each gene (10 sgRNA/gene) relative to sgRNA library plasmid pool.
EGFR inhibitors synergize with anti–PD-1 therapy
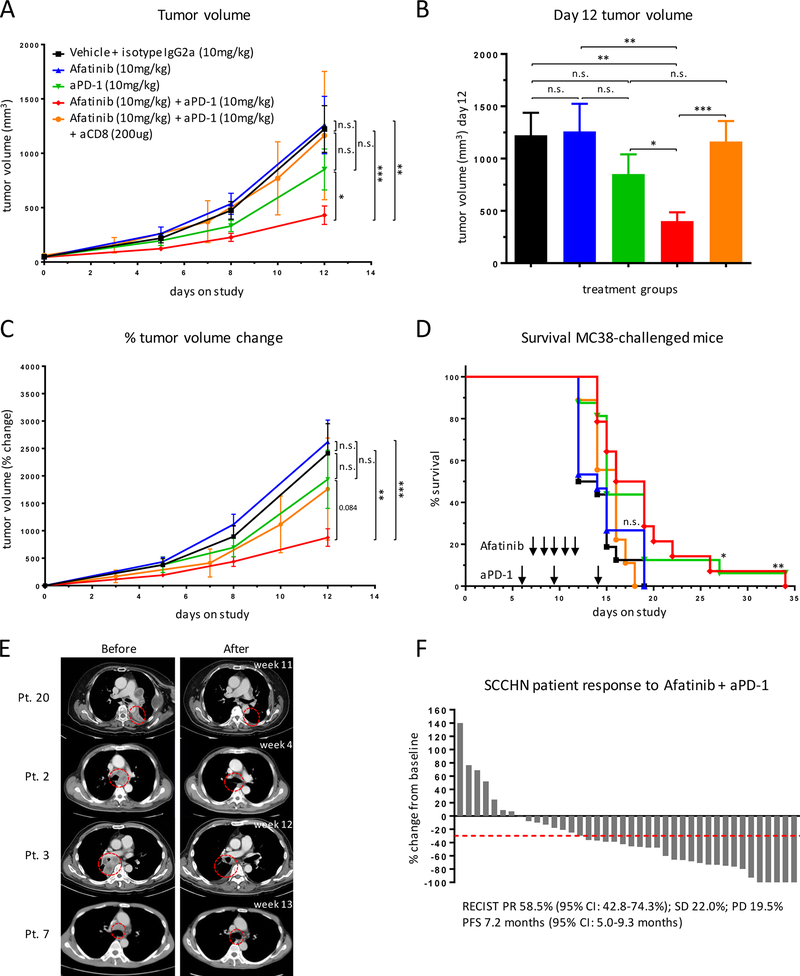
The high antigenicity of the ID8-lucOS model and diffuse nature of the etiology precluded us from using these cells for in vivo validation. We instead utilized the syngeneic MC38 colon due to its well-established, moderate sensitivity to immune checkpoint blockade (23–25). Mice were implanted with MC38 tumors and then treated with vehicle plus isotype control, afatinib, anti–PD-1, or combination afatinib plus anti–PD-1. Combination EGFR inhibition and PD-1 blockade significantly delayed tumor progression relative to vehicle plus isotype control, afatinib, and anti–PD-1 alone (Fig. 7A-C). Combination therapy also conferred significantly improved survival relative to controls (p=0.003), as did anti–PD-1 alone (p=0.026), whereas afatinib single agent had none (p=0.487) (Fig. 7D). Combination afatinib and anti–PD-1 was consistent in its tumor inhibition across all 15 mice, and dosing was well-tolerated (Supplementary Fig. S6). Therapeutic efficacy of the combination treatment was lost when CD8+ T cells were depleted (Fig. 7A-D, orange group), confirming that the effect was immune-mediated. We, therefore, concluded that combination PD-1 blockade and EGFR pharmacological inhibition constitutes a synergistic immunotherapy.
Figure 7: EGFR inhibition enhances efficacy of PD-1 blockade.
C57BL/6J mice (15–16 mice/group) were challenged subcutaneously with 500,000 MC38 colon cancer cells on their flanks and enrolled on-study when tumors reached 50 mm3. Mice were treated with anti-PD-1 on days 5, 8, and 12 and afatinib on days 6, 7, 8, 9, and 10 (where indicated). (A) Tumor volume of enrolled mice treated with the indicated combinations. (B) Tumor volume at day 12 of the study. (C) Tumor growth kinetics. (D) Survival of mice with the indicated treatments. For survival curves: vehicle vs. afatinib n.s.; vehicle vs. anti–PD-1 p=0.013; vehicle vs. combo p=0.003; afatinib vs. anti–PD-1 p=0.069; afatinib vs. combo p=0.017; anti–PD-1 vs. combo n.s.; vehicle vs. combo CD8-depleted n.s.; afatinib vs. combo CD8-depleted n.s.; anti–PD-1 vs. combo CD8-depleted p=0.071; combo vs. combo CD8-depleted p=0.028. (E,F) Response to afatinib and pembrolizumab combination therapy in retrospective cohort of patients with SCCHN. Data presented as (E) pre- and post-treatment scans of selected responders and (F) waterfall plot of % tumor volume change from baseline in response to treatment. Flank tumor growth curves were analyzed using two-way ANOVA, bar graphs were analyzed using unpaired Student’s t-test, and survival experiments used the log-rank Mantel-Cox test for survival analysis. All indicated with *p<0.05, **p<0.01, ***p<0.001.
We compiled a retrospective cohort of 41 relapsed/metastatic squamous-cell carcinoma of the head and neck (SCCHN) patients who received combination afatinib and the PD-1 antibody pembrolizumab (Fig. 7E and F) at the National Taiwan University Hospital between November 2016 and September 2017. Combination therapy resulted in an overall response rate (ORR) of 58.5% by RECIST criteria and an average tumor size reduction of 82.9%, without associated increased toxicity (Fig. 7E and F; Supplementary Table S3). This is compared to reported ORR of 16% to pembrolizumab monotherapy (8) and ORR of 10% to afatinib monotherapy (26) in SCCHN patients. These clinical observations support our preclinical findings demonstrating a synergistic effect of EGFR inhibitors with anti–PD-1 inhibitors.
Discussion
We created a high-throughput screening assay that can be used to identify both drug candidates (plate-based compound screen) and targets (pooled CRISPR/Cas9 screen). Prior studies have paired target cells expressing a model antigen with CD8+ T cells expressing antigen-specific T-cell receptors with the intent to identify tumor cell–intrinsic immunomodulatory genes (27–29). These studies elucidate mechanisms conferring resistance to immune pressure, and our results were concordant, whether from the compound screen (JAK2 inhibitor AZD1480) or CRISPR/Cas9 screen (H2-K1, Tap1, Tap2, and B2m). However, the studies focused on the fundamental biology and specific pathways that tumor cells often mutate or downregulate to evade T-cell recognition and killing, and our work focused on the opposite end: genes that sensitize tumor cells to CD8+ T cell–mediated killing.
EGFR was a logical, if initially unexpected hit. EGFR has previously been shown to antagonize HLA class-I expression via suppression of STAT1 in head and neck cancer patients treated with cetuximab (30). Cetuximab-mediated inhibition of EGFR signaling was associated with enhanced IFNγ receptor 1 (IFNAR) expression, which, through STAT1-dependent signaling, enhanced IFNγ-induced expression of HLA class-I and TAP1/2. In another study, pharmacological inhibitors of EGFR and cetuximab were shown to upregulate basal and IFNγ-induced expression of class I and class II in human keratinocytes. The same study provided in vivo validation where patients already receiving erlotinib or cetuximab consented to skin biopsies, and on-treatment elevation in HLA mRNA was demonstrated (22). A genome-wide CRISPR screen characterizing mechanisms of tumor cell immune evasion has identified SOX10 as a top hit, which conferred resistance to T cell-mediated killing commensurate with B2m, HLA-A, and TAP1 (29). This would plausibly implicate an EGFR-related mechanism, as knockdown of SOX10 in human melanoma was previously shown (31) to result in high expression of EGFR, which dampened antigen processing and presentation, leading to immune escape.
Any modulation of antigen presentation or tumor cell stress is likely to affect NK cell involvement in the antitumor immune response. Pharmacologic inhibition of EGFR with gefitinib or silencing with siRNA increases expression of MHC-I in the PC9 mutEGFR T790M human NSCLC cell line, which is consistent with our data, and downregulates expression of NKG2D ligands MICB and ULBP–2/5/6 (32). Subsequently, gefitinib attenuates NK cell-mediated lysis of tumor cells. In another study, however, EGFR inhibition with gefitinib enhanced NK cell-mediated cytotoxicity of L858R + T790M mutEGFR tumor cells via upregulation of NKG2D ligands MICA, ULBP1, and ULBP2 (33). EGFR inhibition could potentially enhance or inhibit NK cell recognition of tumor cells by modulation of stress ligands recognized by activating NK cell receptors and through KIR-mediated missing-self recognition that is dependent upon expression of MHC class-I (34). It is possible that there are alternative mechanisms of EGFR inhibitor-mediated immunomodulatory function that involves NK cells.
The immunological contribution of oncogenic EGFR has been explored clinically (35) and pre-clinically (36) but mostly as it relates to its regulation of PD-L1 expression in tumor cells. This led to the rational hypothesis that addition of PD-1/PD-L1 blocking antibodies might improve EGFR TKI in EGFR mutant lung cancer by activating the immune infiltrate otherwise suppressed by secondary mechanisms downstream of aberrant EGFR signaling. Therefore, evaluation of EGFR TKIs with anti-PD-1 therapies have mostly been limited to EGFR mutant lung cancer. There are currently two clinical trials exploring this combination: nivolumab plus EGF816 (NCT02323126) and nivolumab plus erlotinib (NCT01454102). A trial of osimertinib, a mutant selective EGFR inhibitor, combined with the PD-L1 inhibitor durvalumab in patients with T790M EGFR mutant lung cancer was stopped due to toxicity (NCT02454933). EGFR TKIs are the standard of care for lung cancers harboring EGFR activating mutations but exhibit minimal therapeutic efficacy in wild-type EGFR NSCLC (37,38), colorectal cancer (39), and SCCHN (26). Only afatinib has an approval in a wild-type EGFR setting. Yet, EGFR mutant lung cancer, the largest cohort of patients treated with EGFR inhibitors, may not be an ideal setting in which positive immunomodulatory properties would necessarily be noticed, largely due to the immunologically “cold” nature of the disease, as our group and others have shown (40). The data presented above confirming EGFR as an immune-oncology target was conducted in three distinct wild-type EGFR models. This suggests that, whether through its regulation of PD-L1 or basal and IFNγ-induced antigen processing and presentation, inhibition of EGFR may be broadly efficacious across mutant EGFR and wild-type EGFR cancers. The oncogenic properties of EGFR are well-established, but our work supports the classification of EGFR as an immune-oncology target. Given the FDA-approval of multiple pharmacologic and biologic inhibitors of EGFR and their established clinical application, inclusion of inhibitors to non-mutated EGFR is an attractive approach to amplify the immunogenicity of tumors treated with immune checkpoint blockade. Our human data in SCCHN is evidence for the utility of this approach. The retrospective clinical data, combined with the synergistic effect of afatinib and anti–PD-1 in our in vivo model, suggested that the combination of immune checkpoint blockade with EGFR TKI may have a therapeutic benefit in wild-type EGFR tumors. Although our clinical analysis was conducted retrospectively, the findings suggest that EGFR TKIs and anti–PD-1 agents should be evaluated in prospective clinical trials, specifically in wild-type EGFR cancers.
EGFR TKI trials report high incidences of adverse events, such as skin rashes, in up to 66–90% of patients (37–39,41). There is ample evidence both in this paper and in published literature to support the assertion that these drugs induce upregulation of MHC class-I. This could potentially cause aberrant T-cell recognition of self-antigen. Breaking of tolerance may explain the high rates of toxicity observed with EGFR TKI, and they may be immune-mediated. Immune activation may also explain the therapeutic benefit observed in wild-type EGFR lung and colorectal patients treated with EGFR TKI and, consistent with this this hypothesis, the benefit of EGFR TKIs is often greater in patients who develop a skin rash (37,38). Adverse events are likely to remain consistent, if not become exacerbated, by combination with immune checkpoint blockade. In our limited dataset of 41 patients, we did not observe this compounded toxicity with combination therapy. However, this was a retrospective analysis and not a prospective clinical trial. If in future combination trials toxicity is limiting, this could be potentially mitigated by using EGFR inhibitors at lower dosages or by evaluating different treatment schedules, including sequencing or intermittent dosing of EGFR TKIs.
We present here an assay for high throughput screening that can be utilized to identify novel immunomodulatory therapeutics and current drugs that would logically be expected to augment immune checkpoint blockade and other developing immunotherapies. As currently designed, one OT-I mouse spleen with a routine harvest of 10–12 million CD8+ T cells is sufficient to plate 10–12 96-well assay plates, rendering analysis of compound libraries in the hundreds to thousands feasible. Our initial screen identified EGFR as a target that sensitizes tumor cells to CD8+ T cell–mediated killing, a result which was confirmed in two different murine tumor cell lines and independently validated in a pooled CRISPR/Cas9 screen. Inhibition of EGFR with afatinib enhanced anti–PD-1 therapeutic efficacy in vivo in the MC38 syngeneic colon cancer model and in human SCCHN patients. The CTL OT-I assay is a tool to rationally identify promising drug combinations to enhance immunotherapy, which is rapidly becoming a cornerstone of medical oncology.
Supplementary Material
Acknowledgements
We thank the patients and their families for agreeing to follow-up study and for granting us access to their medical records. We thank the Robert A. and Renée E. Belfer Foundation, Expect Miracles Foundation, and American Cancer Society (grant #CRP-17–111-01-CDD) for their generous support. We thank Harvard Medical School for access to their LINCS kinase library (grant #NIH U54 HL127365).
This work was generously supported by the Robert A. and Renée E. Belfer Foundation, Expect Miracles Foundation, and ACS grant #CRP-17–111-01-CDD. LINCS kinase library was made available through grant number NIH U54 HL127365.
P.A.J. receives commercial research support from AstraZeneca and Boehringer Ingelheim, and provides consulting for AstraZeneca, Boehringer Ingelheim, Pfizer and Roche/Genentech
References
- 1.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, et al. Nivolumab and Ipilimumab versus Ipilimumab in Untreated Melanoma. N Engl J Med. 2015;372:2006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall Survival and Long-Term Safety of Nivolumab (Anti–Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non–Small-Cell Lung Cancer. J Clin Oncol. 2015;33:2004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferris RL, Blumenschein GJ, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375:1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, et al. Atezolizumab as first-line therapy in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet Lond Engl. 2017;389:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015;373:1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Younes A, Santoro A, Shipp M, Zinzani PL, Timmerman JM, Ansell S, et al. Nivolumab for classical Hodgkin’s lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: a multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 2016;17:1283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon ED, Drake CG, Scher HI, Fizazi K, Bossi A, Eertwegh AJM van den, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184–043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch TJ, Bondarenko I, Luft A, Serwatowski P, Barlesi F, Chacko R, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter phase II study. J Clin Oncol Off J Am Soc Clin Oncol. 2012;30:2046–54. [DOI] [PubMed] [Google Scholar]
- 11.Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, et al. Ipilimumab plus Dacarbazine for Previously Untreated Metastatic Melanoma. N Engl J Med. 2011;364:2517–26. [DOI] [PubMed] [Google Scholar]
- 12.DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, et al. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu C, Wehr DR, Edwards J, Hauge B. Rapid one-step recombinational cloning. Nucleic Acids Res. 2008;36:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, et al. The Myeloma Drug Lenalidomide Promotes the Cereblon-Dependent Destruction of Ikaros Proteins. Science. 2014;343:305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The Differential Effects of Mutant p53 Alleles on Advanced Murine Lung Cancer. Cancer Res. 2005;65:10280–8. [DOI] [PubMed] [Google Scholar]
- 17.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. [DOI] [PubMed] [Google Scholar]
- 18.Schulz RA, Yutzey KE. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev Biol. 2004;266:1–16. [DOI] [PubMed] [Google Scholar]
- 19.Kwiatkowski N, Deng X, Wang J, Tan L, Villa F, Santaguida S, et al. Selective Aurora Kinase Inhibitors Identified Using a Taxol-Induced Checkpoint Sensitivity Screen. ACS Chem Biol. 2012;7:185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miduturu CV, Deng X, Kwiatkowski N, Yang W, Brault L, Filippakopoulos P, et al. High-Throughput Kinase Profiling: A More Efficient Approach toward the Discovery of New Kinase Inhibitors. Chem Biol. 2011;18:868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cross DAE, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollack BP, Sapkota B, Cartee TV. Epidermal Growth Factor Receptor Inhibition Augments the Expression of MHC Class I and II Genes. Clin Cancer Res. 2011;17:4400–13. [DOI] [PubMed] [Google Scholar]
- 23.Deng L, Liang H, Burnette B, Beckett M, Darga T, Weichselbaum RR, et al. Irradiation and anti–PD-L1 treatment synergistically promote antitumor immunity in mice. J Clin Invest. 2014;124:687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngiow SF, Young A, Jacquelot N, Yamazaki T, Enot D, Zitvogel L, et al. A Threshold Level of Intratumor CD8+ T-cell PD1 Expression Dictates Therapeutic Response to Anti-PD1. Cancer Res. 2015;75:3800–11. [DOI] [PubMed] [Google Scholar]
- 25.Zippelius A, Schreiner J, Herzig P, Müller P. Induced PD-L1 Expression Mediates Acquired Resistance to Agonistic Anti-CD40 Treatment. Cancer Immunol Res. 2015;3:236–44. [DOI] [PubMed] [Google Scholar]
- 26.Clement PM, Gauler T, Machiels JP, Haddad RI, Fayette J, Licitra LF, et al. Afatinib versus methotrexate in older patients with second-line recurrent and/or metastatic head and neck squamous cell carcinoma: subgroup analysis of the LUX-Head & Neck 1 trial. Ann Oncol. 2016;27:1585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan D, Kobayashi A, Jiang P, Andrade LF de, Tay RE, Luoma A, et al. A major chromatin regulator determines resistance of tumor cells to T cell–mediated killing. Science. 2018;eaao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava RM, Trivedi S, Concha-Benavente F, Hyun-bae J, Wang L, Seethala RR, et al. STAT1-Induced HLA Class I Upregulation Enhances Immunogenicity and Clinical Response to Anti-EGFR mAb Cetuximab Therapy in HNC Patients. Cancer Immunol Res. 2015;3:936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C, Wang L, Huang S, Heynen GJJE, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22. [DOI] [PubMed] [Google Scholar]
- 32.Okita R, Wolf D, Yasuda K, Maeda A, Yukawa T, Saisho S, et al. Contrasting Effects of the Cytotoxic Anticancer Drug Gemcitabine and the EGFR Tyrosine Kinase Inhibitor Gefitinib on NK Cell-Mediated Cytotoxicity via Regulation of NKG2D Ligand in Non-Small-Cell Lung Cancer Cells. PLoS ONE [Internet]. 2015;10 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4595469/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He S, Yin T, Li D, Gao X, Wan Y, Ma X, et al. Enhanced interaction between natural killer cells and lung cancer cells: involvement in gefitinib-mediated immunoregulation. J Transl Med. 2013;11:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16:7–19. [DOI] [PubMed] [Google Scholar]
- 35.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2015;10:910–23. [DOI] [PubMed] [Google Scholar]
- 36.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 Pathway Contributes to Immune Escape in EGFR-Driven Lung Tumors. Cancer Discov. 2013;3:1355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
- 38.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–32. [DOI] [PubMed] [Google Scholar]
- 39.Townsley CA, Major P, Siu LL, Dancey J, Chen E, Pond GR, et al. Phase II study of erlotinib (OSI-774) in patients with metastatic colorectal cancer. Br J Cancer. 2006;94:1136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lizotte PH, Ivanova EV, Awad MM, Jones RE, Keogh L, Liu H, et al. Multiparametric profiling of non–small-cell lung cancers reveals distinct immunophenotypes. JCI Insight [Internet]. 2016. [cited 2018 Feb 7];1 Available from: https://insight-jci-org.ezp-prod1.hul.harvard.edu/articles/view/89014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merla A, Goel S. Novel Drugs Targeting the Epidermal Growth Factor Receptor and Its Downstream Pathways in the Treatment of Colorectal Cancer: A Systematic Review. Chemother Res Pract [Internet]. 2012;2012 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3477664/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.