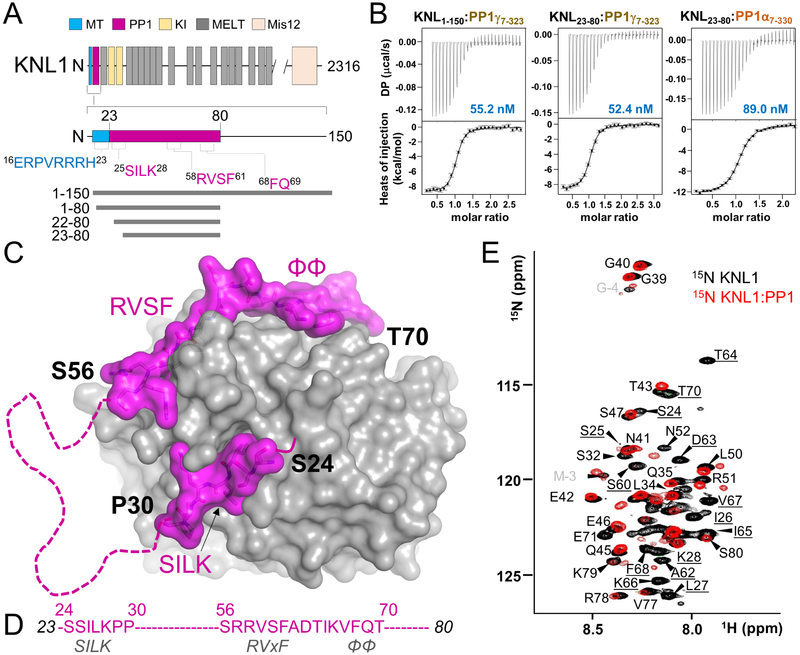
Figure 1: KNL1 binds PP1 using three PP1-specific SLiMs separated by a disordered loop.
(A) KNL1 domain structure, highlighting the putative protein interaction sites (microtubule, MT, blue; PP1, magenta). The PP1 binding domain is hypothesized to contain three established PP1 binding SLiMs, namely the SILK, RVxF and ΦΦ motifs, while the putative MT binding domain is positively charged. The constructs and variants examined in this study are shown below.
(B) Binding isotherms of KNL1 variants with either PP1γ or PP1α.
(C) Crystal structure of the KNL123–80:PP1 holoenzyme. PP1 is in grey and KNL1 in magenta. Residues not modeled due to a lack of density are indicated by a dashed line.
(D) The ordered KNL1 residues are specified in text (magenta). The established PP1 binding motifs present in KNL1 are indicated in grey text.
(E) Overlay of the 2D [1H,15N] HSQC spectra of KNL123–80 (black) with the KNL23–80:PP1 holoenzyme (red). Peaks observed in the KNL23–80:PP1 holoenzyme spectrum correspond to residues that remain disordered upon complex formation. Peaks present only in the KNL123–80 spectrum are underlined.
Related to Figures S1–S3