Abstract
Objective
Brown adipose tissue (BAT) is important for thermoregulation in many mammals. Uncoupling protein 1 (UCP1) is the critical regulator of thermogenesis in BAT. Here we aimed to investigate the deacetylation control of BAT and to investigate a possible functional connection between UCP1 and sirtuin 3 (SIRT3), the master mitochondrial lysine deacetylase.
Methods
We carried out physiological, molecular, and proteomic analyses of BAT from wild-type and Sirt3KO mice when BAT is activated. Mice were either cold exposed for 2 days or were injected with the β3-adrenergic agonist, CL316,243 (1 mg/kg; i.p.). Mutagenesis studies were conducted in a cellular model to assess the impact of acetylation lysine sites on UCP1 function. Cardiac punctures were collected for proteomic analysis of blood acylcarnitines. Isolated mitochondria were used for functional analysis of OXPHOS proteins.
Results
Our findings showed that SIRT3 absence in mice resulted in impaired BAT lipid use, whole body thermoregulation, and respiration in BAT mitochondria, without affecting UCP1 expression. Acetylome profiling of BAT mitochondria revealed that SIRT3 regulates acetylation status of many BAT mitochondrial proteins including UCP1 and crucial upstream proteins. Mutagenesis work in cells suggested that UCP1 activity was independent of direct SIRT3-regulated lysine acetylation. However, SIRT3 impacted BAT mitochondrial proteins activities of acylcarnitine metabolism and specific electron transport chain complexes, CI and CII.
Conclusions
Our data highlight that SIRT3 likely controls BAT thermogenesis indirectly by targeting pathways upstream of UCP1.
Keywords: Brown adipose tissue, Thermogenesis, Thermoregulation, Mitochondria, Uncoupling protein 1, Sirtuin 3, Acetylation, Oxidative phosphorylation, Fatty acid oxidation, Acylcarnitines
Highlights
-
•
Impaired cold tolerance in Sirt3KO mice under fed conditions.
-
•
Defective UCP1 dependent respiration in BAT mitochondria of Sirt3KO mice.
-
•
SIRT3 and cold stress reshape the mitochondrial acetylome of BAT mitochondria.
-
•
Activation of BAT in Sirt3KO mice results in changes to plasma acylcarnitines.
-
•
Individual ETC complexes are defective in BAT mitochondria in absence of SIRT3.
1. Introduction
Brown adipose tissue (BAT) helps many mammals, including humans, maintain body temperature in cold environments through a process called non-shivering thermogenesis [1], [2], [3], [4]. BAT thermogenesis can be stimulated by cold exposure and by pharmacological administration of adrenergic receptor agonists [5]. BAT activation results in the uptake and oxidation of large amounts of glucose and fatty acids (FAs) in a process that can markedly increase whole body energy expenditure in mice [6], [7].
BAT thermogenesis is dependent on the activity of uncoupling protein-1 (UCP1), an abundant protein in the mitochondrial inner membrane that induces proton leak-mediated respiration [8], [9]. When BAT is activated, Ucp1 gene expression is increased by the action of multiple transcription factors including the peroxisome proliferator activated receptor (PPAR) and CCAAT/enhancer binding protein (C/EBP) families and cyclic AMP (cAMP) response binding protein (CREB), as well as thyroid hormone [10], [11]. These transcription factors are activated by p38 mitogen-activated protein kinase (MAPK), which is considered as the central mediator of the cAMP signaling pathway that induces brown adipocyte thermogenesis [12]. UCP1-mediated thermogenesis falls under the regulation of the sympathetic nervous system via the activation of adrenergic receptors, and subsequently stimulation of lipolysis in BAT. The liberated FAs are then used for the allosteric activation of UCP1, and to support greatly increased rates of mitochondrial fatty acid oxidation (FAO). The latter provides reducing equivalents/electrons to drive the electron transport chain (ETC), which establishes the inner membrane protonmotive force that in turn drives the protonophore activity of UCP1 [5], [13], [14].
BAT has high expression of sirtuin 3 (SIRT3), which is an NAD+-dependent deacetylase in the mitochondria [15]. A previous study showed that cold increases the transcription of SIRT3 mRNA in BAT; however the impact on protein levels is unclear [16]. SIRT3 is the master regulator of deacetylation in the mitochondria [15] while the other mitochondrial sirtuins, SIRT4 and SIRT5 do not have significant roles in mitochondrial deacetylation [15], [17], [18], [19]. Large-scale acetylome analyses from multiple studies demonstrate that mitochondrial proteins are frequently targets for lysine acetylation [20], [21], [22], [23]. While lysine acetylation outside of the mitochondria is catalyzed by lysine acetyltransferase (KAT) enzymes, no mitochondrial KAT enzyme in BAT has been described to date. Overall mitochondrial acetylation levels are correlated with the level of acetyl-CoA [24], and much of the acetylation that occurs in mitochondria is thought to occur non-enzymatically, owing in part to the high concentrations of acetyl-CoA as a product of FAO [25]. SIRT3 appears to play a critical role in reversing these such non-enzymatic acetylation, which are generally thought to have an inhibitory effect on enzymatic activities in mitochondria [26]. Consistent with this, Hirschey et al. showed that the absence of SIRT3 in fasting mice is associated with decreased FAO rates in various tissues, including BAT, and these mice are intolerant to acute cold stress [21]. Altogether these findings are consistent with the notion that SIRT3 could be a critical regulator of BAT thermogenesis, and that the underlying molecular mechanisms merit investigation.
To investigate the control of SIRT3-mediated deacetylation of BAT thermogenesis, we studied the metabolic phenotypes of BAT from Sirt3 knock-out (Sirt3KO) mice that were cold exposed for 2 days, or were held at thermoneutrality and injected with the β3-adrenergic agonist CL316,243. We report that mice deficient in SIRT3 display widespread defects in BAT lipid use/oxidation and in thermoregulation even in fed conditions. We also show that respiration is decreased in mitochondria isolated from BAT of Sirt3KO mice relative to wild-type controls. While SIRT3 impacted acetylation sites on UCP1 in room temperature housed mice, there were little SIRT3-dependent effects at these same sites after cold stress and mutation of these sites to prevent acetylation did not affect protein function in a cellular model. In contrast, the absence of SIRT3 impacted acetylation on diverse FAO/acylcarnitine-related enzymes and ETC proteins in BAT mitochondria under both conditions. Functions controlled by these proteins are defective in Sirt3KO mice under conditions that activate BAT in vivo. Together, our findings suggest that SIRT3 indirectly controls BAT thermogenesis by deacetylating and promoting the function of pathways upstream of UCP1.
2. Materials and methods
2.1. Animals
Sirt3 whole body knock-out (KO) mice were described previously [27]. Sirt3 knock-out was confirmed by PCR before experimentation. Mice were given ad libitum, a standard rodent chow (44.2% carbohydrates, 6.2% fat, and 18.6% crude protein; diet T.2018, Harlan Teklad, Indianapolis IN) and water, and were housed with a 12/12 h light–dark cycle in ventilated cages. At an age of 5–8 weeks, male mice were used in two different experimental approaches to maximize the activity of BAT. Mice were kept either at room temperature (24 °C), housed in groups of 2–3 mice, or were exposed to 4 °C for 2 days, housed individually, with free access to water and food. Body weights were taken, as well as the weights of epididymal white adipose tissue (eWAT) and interscapular BAT (iBAT) after sacrificing mice by cervical dislocation. Tissues were used immediately for experiments or were flash frozen and stored at −80 °C for later analyses. In other experiments, mice were injected with the β3-adrenergic agonist, CL 316,243 (1 mg/kg i.p.) and were housed at 28 °C. All animal experimental procedures were in accordance with the guidelines and principles of the Canadian Council of Animal Care and after the approval of the Animal Care Committee of the University of Ottawa.
2.2. Body composition and comprehensive metabolic phenotyping
EchoMRI (EchoMRI, Houston TX) was used to assess lean body mass, and fat mass of mice. To examine characteristics of metabolic phenotype we used a Comprehensive Lab Animal Monitoring System (CLAMS; Columbus Systems, Columbus OH). During CLAMS analyses, mice were housed individually at 28 °C for at least 2 days for acclimation purposes and then were exposed to 4 °C for two days in the CLAMS system. In other experiments to address BAT-specific thermogenic activity, mice were studied at thermoneutrality (28 °C) before and after CL 316,243 injections. Mice were fasted 3 h prior to, and following the 10:00 bolus i.p. injection of CL 316,243 (1 mg/kg) to avoid food associated effects (e.g., foraging activity; thermic effect of food). Pre-CL injection values were averaged (07:00–10:00). Post-CL injection values were the average of the value at 90-minute post-injection, and the value immediately preceding that one.
2.3. Histological analyses of BAT
iBAT from mice that were housed at room temperature or exposed to the cold was dissected, cleaned to remove any white adipose, muscle or connective tissues, and then fixed in 10% formalin overnight. iBAT was then ethanol dehydrated and stored in 70% ethanol prior to paraffin embedding. Paraffin embedded tissue was sectioned to the largest surface area and used for Hematoxylin and Eosin (H&E) staining. After H&E staining, Mirax Viewer Image software (version 1.6) was used to analyze the sections in a ZEISS-MIRAX Midi Slide scanning system (Zeiss Microimaging, Oberkochen, Germany, and 3DTech, Budapest, Hungary) and digital images were acquired at 20x magnification. Images were extracted using Aperio ImageScope software (version 12.3.3; Leica Biosystems), and the extracted images were analyzed using FIJI (ImageJ; NIH) software [28]. The range thresholding that was applied across all replicate images in all conditions was (200–255). Finally, the total image area was measured, and a percent lipid area was calculated.
2.4. Core body temperature measurement
The core body temperature of mice was measured using an anal thermometer (Physitemp TH-8 Thermalert Monitoring Thermometer, Physitemp, USA) before and after cold exposure.
2.5. BAT mitochondrial isolation
Mitochondria were isolated from iBAT of wild-type (WT) and Sirt3KO mice as described previously [29]. Briefly, on the day of the experiment, iBAT of 1–3 male mice from each genotype, was quickly dissected, cleaned and kept on ice. iBAT was then rapidly cut with scissors into small pieces, before being homogenizing in 0.25M sucrose using 4 to 5 strokes in a Potter Elvehjem teflon–glass tissue grinder. The iBAT homogenate was subjected to differential centrifugation steps starting with a spin at 8500g for 10 min, to remove the fat layers. After removing the supernatant, the wall of the tubes was wiped to remove any residual amount of fat following by re-suspending pellets in the sucrose buffer (0.25M). To remove nuclei and cell debris, the suspension was then spun at 800g for 10 min. The mitochondria-containing supernatants were collected and were spun at 8500g for 10 min to pellet the mitochondria. Finally, the mitochondria were washed with 0.25M sucrose containing 0.3% fatty acid-free bovine serum albumin for 10 min at 8500g to remove endogenous fatty acids. The mitochondrial pellet was gently re-suspended in specific buffers depending on the downstream applications (see below).
2.6. Western blotting
Mitochondrial pellets were resuspended in 0.25M sucrose supplemented with 10 mM sodium butyrate (Sigma Aldrich, #303410), 10 mM nicotinamide (Sigma Aldrich, #N3375) and protease inhibitor mixture (Complete Mini, Roche). Protein concentration was quantified using the Bradford method (Biorad). 10 μg of mitochondrial proteins was loaded per lane onto a 12% SDS-polyacrylamide gel. After electrophoresis, proteins were transferred by electro-blotting to nitrocellulose membrane. Nitrocellulose membrane was used for all blots and was blocked with 5% BSA for 1 h. After that, membranes were incubated with primary antibodies to examine the expression of SIRT3, UCP1, total mitochondrial acetylation levels, and OXPHOS complexes. Western blotting was performed with the following antibodies: anti-SIRT3 antibody (1: 1,500; Cell Signaling, #5490S); anti-UCP-1 antibody (1: 3,000; Sigma Aldrich, #U6382); anti-acetylated lysine antibody (1:1500; Calbiochem # ST1027); total OXPHOS rodent antibody cocktail (1:2000; Abcam, #Ab110413); and anti-cytochrome c oxidase antibody (1: 4,000; Invitrogen #459600). Cytochrome c oxidase was used as a mitochondrial loading control. Western blots of UCP1 and total mitochondrial acetylation in Figure 2, Figure 3C were probed from the same membrane. Quantification was performed by using the ImageJ software (NIH).
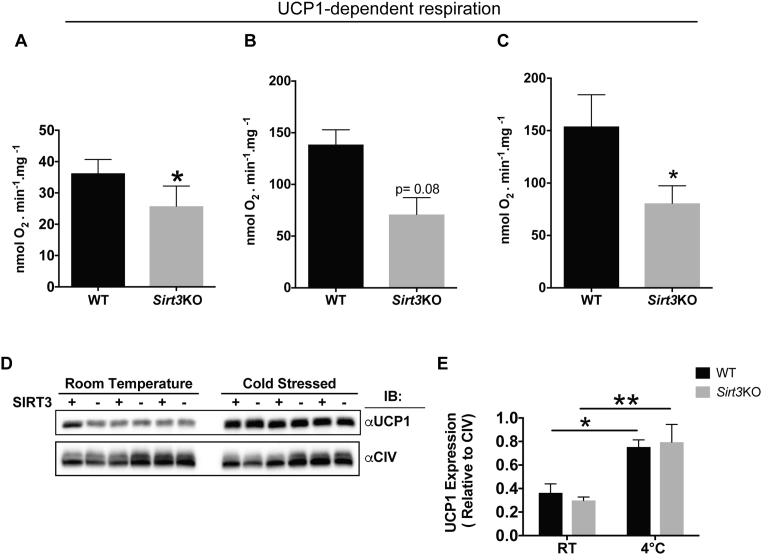
Figure 2.
UCP1 dependent respiration is decreased in BAT of Sirt3KO mice. (A) UCP1 dependent respiration was measured in isolated mitochondria from iBAT of room temperature housed WT and Sirt3KO mice. 0.35 mg Mitochondria were energized with 25 μM PLC and UCP1 dependent respiration was determined as that which was inhibited by 2 mM GDP. N = 6/group. (B) UCP1 dependent respiration was measured in isolated mitochondria from iBAT of room temperature housed WT and Sirt3KO mice. 0.25 mg Mitochondria were energized with 5 mM G3P and UCP1 dependent respiration was determined as that which was inhibited by 2 mM GDP. N = 4/group. (C) UCP1 dependent respiration was measured in isolated mitochondria from iBAT of cold exposed WT and Sirt3KO mice. 0.25 mg Mitochondria were energized with 5 mM G3P and UCP1 dependent respiration was determined as that inhibited by 2 mM GDP. N = 4/group. (D) Western blotting analysis of UCP1 expression. N = 3/group. (E) Quantification of UCP1 western blotting. Data are represented as mean ± SEM. Student's t-test; Two-tailed and two-way ANOVA with Sidak's test were used, *p < 0.05 and **p < 0.01.
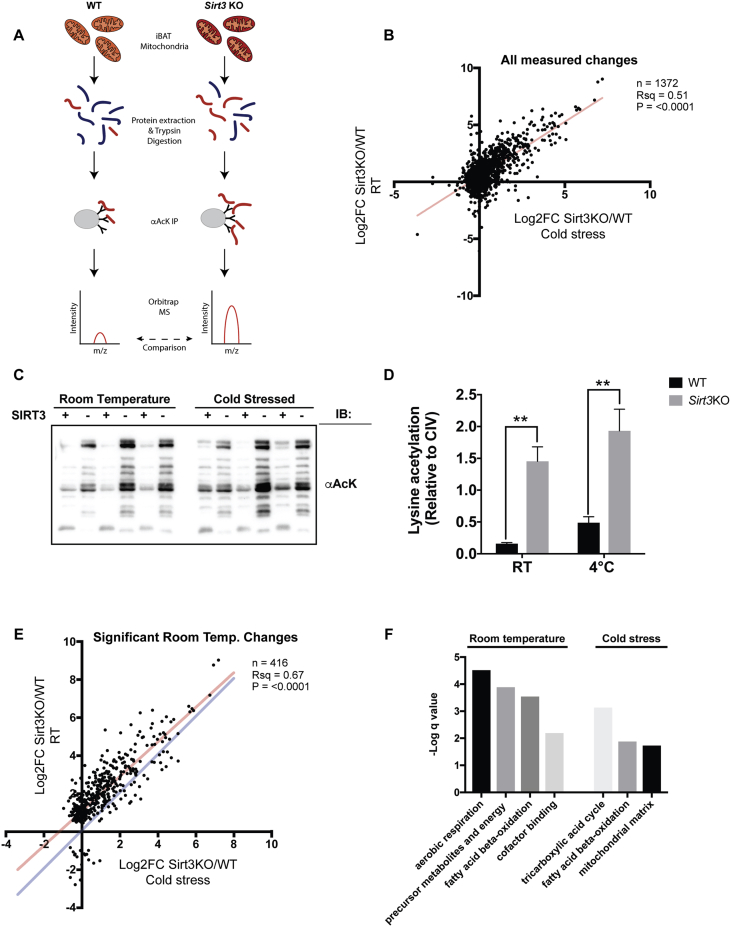
Figure 3.
Cold and the absence of SIRT3 increase mitochondrial acetylation. (A) Schematic diagram of acetylome profiling of isolated BAT mitochondria. Proteins were extracted from iBAT mitochondria under denaturing conditions prior to trypsinization and immunopurification with anti-acetyllysine antibodies and analysis using Orbitrap MS, as described in the experimental procedures. Two distinct experiments were carried out comparing wild-type to Sirt3KO animals, housed at either room temperature (experiment 1) or at 4 °C for 2 days (experiment 2). N = 6 BAT mitochondria samples for each genotype. (B) Measured Log2FC Sirt3KO/WT was plotted for acetylated lysines detected via MS for room temperature versus cold treated mice. Log2FC computed using MSStats, as described in the experimental procedures. Linear regression calculated using GraphPad Prism. This analysis includes significant and non-significant changes. (C) Total mitochondrial acetylation was assessed by western blotting from samples described in Figure 2D. N = 3/group. Note: same blots as Figure 2D were probed for mitochondrial acetylation. As such, the loading control of the acetylation blot is the same loading control in as Figure 2D. (D) Quantification of mitochondrial acetylation western blotting. Data are represented as mean ± SEM. Two-way ANOVA with Sidak's test was used, **p < 0.01. (E) Greater measured impact of SIRT3 at room temperature relative to cold stressed animals. Measured Log2FC Sirt3KO/WT was plotted for acetylated lysines detected via MS for room temperature versus cold treated mice. Log2FC computed using MSStats, as described in the experimental procedures. Linear regression calculated using GraphPad Prism. Plotted are significantly-regulated sites detected in samples from room temperature housed animals, regardless of fold-change versus corresponding fold-change detected in cold stressed animals, regardless of significance. Red line indicates the best fit line of linear regression. The blue line represents slope = 1. The majority of data points above blue line suggests more dramatic changes in room temperature housed animals. The graph contains a subset of the data plotted in Figure 3B. (F) GO-term enrichment of SIRT3-regulated lysine acetylation sites calculated using Metascape for room-temperature and cold-regulated sites (log2 Sirt3KO/WT > 1; adjusted p < 0.05). Only sites for which numerical p values could be calculated were used for this analysis. q values represent p values of GO-term analyses following adjustment for multiple-testing.
2.7. Label free mass spectrometry acetylome profiling
Sample preparation for acetylome profiling followed a protocol originally developed for yeast Saccharomyces cerevisiae [30], with appropriate modifications implemented for mouse BAT. For each treatment group (warm or cold), all samples were processed at the same time to ensure consistency. N = 6 for each treatment group, where a single biological replicate is one mitochondrial protein prep from iBAT isolated from one mouse. Generation of AcK peptides: Mitochondrial protein samples were suspended in 1 mL of AcK buffer (8M Urea, 0.1 M Tris–HCl pH 8.0, 150 mM NaCl, 10 mM sodium butyrate (Sigma Aldrich 303,410), 10 mM nicotinamide (Sigma Aldrich 3376), with Roche protease inhibitor tablet (w/o EDTA). Resuspended samples were quantitated using a BCA assay. For each sample, 0.4 or 1.4 mg of mitochondrial protein from iBAT of room temperature housed or cold exposed mice, respectively, was reduced using TCEP (Sigma Aldrich C4706–2, 4 mM final concentration) prior to alkylation with iodoacetamide (Sigma Aldrich L1149–5G, 10 mM final concentration), and quenching with DTT (10 mM final concentration). Subsequently, samples were diluted to < 2M urea with 0.1 M Tris–HCl pH 8.0 and digested with trypsin (Pierce 90,058, ∼15 μg/sample). Digestions were performed at room temperature overnight. Samples were acidified using 10% TFA to a pH of <3, which was monitored by spotting on pH paper. Samples were clarified by centrifugation at 10,000 g at room temperature to remove precipitate and loaded onto a Sep Pak tC18 column (Waters). Column activation was with 1 ml 80% ACN/0.1% TFA. Equilibration was with 3 × 1 ml 0.1% TFA. The columns were washed with 5 × 1 ml 0.1% TFA. Bound peptides were eluted with 1 ml 40% ACN/0.1% TFA. AcK enrichment: The eluted peptides were frozen and lyophilized prior to resuspension in AcK IP buffer (0.1 M Tris–HCl pH 8, 50 mM NaCl, Roche protease inhibitor tablet w/o EDTA) and clarification by centrifugation (2 min at 10,000 rpm). Supernatant was incubated with 50 μL packed volume of ImmuneChem anti-AcK agarose beads (ICP0388) at 4 °C for 2 h. Beads were washed twice with 1 mL lysis buffer and twice with 1 mL water. Two elutions of 60 μL of 0.15% TFA (10 min) were pooled. Peptides were purified on Pierce C18 spin tips (84,850) according to manufacturer's instructions. Elutions were dried on a speed vac.
Mass spectrometry analysis: Mass spectrometry analysis was carried out on a Thermo Fisher Orbitrap Fusion with an Easy nLC 1200 ultra-high pressure liquid chromatography system interfaced via a Nanospray Flex nanoelectrospray source. Peptide samples were injected on a C18 reverse phase column (25 cm x 75 um packed with ReprosilPur C18 AQ 1.9 um particles). Peptides were separated via an organic gradient from 5% to 30% ACN in 0.1% formic acid (112 min, flow rate of 300 nl/min). All samples were analyzed in technical duplicate. Data acquisition: The mass spectrometer acquired the spectra in a data-dependent manner throughout the gradient, acquiring a full scan in the Orbitrap (at 120,000 resolutions with an AGC target of 200,000 and a maximum injection time of 100 ms). This was followed by as many MS/MS scans as could be acquired on the most abundant ions in 3s in the dual linear ion trap (rapid scan type, intensity threshold of 5000, HCD collision energy of 29%, AGC target of 10,000, a maximum injection time of 35 ms, and an isolation width of 1.6 m/z). For this dataset, singly and unassigned charge states were rejected. Dynamic exclusion was enabled with a repeat count of 1, an exclusion duration of 20 s, and an exclusion mass width of +/− 10 ppm. Extraction: Raw mass spectrometry data were assigned to murine protein sequences. MS1 intensities extracted with the MaxQuant software package (version 1.5.5.1) [31]. Data were searched against the SwissProt murine protein database (downloaded on January 11, 2016). In addition to lysine acetylation, variable modifications were allowed for N-terminal protein acetylation, methionine oxidation. A static modification was used for carbamidomethyl cysteine. Data analysis: The MaxQuant data were analyzed using our in-house computational pipeline for statistical analysis of relative quantification with fixed and/or mixed effect models, implemented in the MSstats Bioconductor package (version 2) [32]. Contaminants, decoy hits, and peptides not containing acetyllysine residues were removed. Samples for each comparison (WT versus Sirt3KO in either warm or cold conditions) were normalized across fractions by median-centering the log2-transformed MS1-intensity distributions. MS stats group comparison functions were run with the following settings: no interaction terms for missing values, no interference, unequal intensity feature variance, restricted technical and biological scope of replication. Statistically significant changing sites between wild-type and Sirt3KO mice were selected by applying a log2-fold-change (>1.0) and an adjusted p-value (<0.05) corrected for multiple testing threshold. Warm and cold treatment conditions were analyzed separately since these groups were analyzed at different times. Peptides identified by MaxQuant that did not contain KAc sites were extracted from the data to compare KAc site changes to underlying protein abundance changes. The data were analyzed by MSstats in an identical fashion as the KAc site analysis. Protein L2FC values were normalized by median-centering to account for systematic sample loading errors and then were plotted against KAc site L2FC values where both protein and site values could be calculated. GO term enrichment: Analysis was performed using the Metascape analysis tool [33]. Proteins with KAc sites with L2FC Sirt3KO/WT > 1.0 or L2FC and with adjusted p-value < 0.05 were extracted as the gene set for enrichment. A custom background gene set was set as the full list of proteins detected in each experiment. The top terms in each summary group are shown.
2.8. Plasmid design
Synthetic murine UCP1 constructs were ordered from ATUM (https://www.atum.bio). UCP1(WT) encodes the sequence dictated by Uniprot accession P12242. UCP1(K−R) and UCP1(K−Q) constructs contain mutations coding for appropriate changes to lysines 56 and 151. Constructs are expressed from the CMV promoter in vector pD2610-v10. The GFP control vector was also purchased from ATUM and encodes GFP under the control of the same vector, with the designation of pD2610-v10-03.
2.9. Culture and transfection of HEK293T cells
HEK293T cells were cultured in high glucose DMEM medium (Gibco, ThermoFisher) provided with 10% FBS (Wisent) and 1% Antibiotics-Antimycotic (Gibco, ThermoFisher). HEK293T cells were passaged when they reached 90% confluency. For transfection, 70,000 cells were seeded per well in 24-well plates and left overnight. 24 h after seeding, cells were transfected with the different plasmids (500ng/well) as described in the manufacturer's protocol (ThermoFisher, #15338100). After 8 h of transfection, the cells were trypsinized and their viability was measured to be greater that 90% using Trypan blue staining. Transfected cells were seeded in 96-well Seahorse plates or 24-well dishes for western blot confirmation of UCP1 expression. After 24h, cells were analyzed using a Seahorse 96e XF Analyzer and a modified mitochondrial stress assay. The latter assay was conducted as follows.
2.10. Seahorse analysis of UCP1-mediated leak respiration in transfected cells
On the day of the assay, growth medium was changed to Seahorse medium for 1 h prior to the actual analyses; this medium consisted of phenol-free DMEM, including 10 mM glucose, 10 mM pyruvate, and 0.4% BSA. The mitochondrial stress test protocol was modified to examine UCP1-mediated proton leak, and the following reagents were used: (4-[(E)-2-(5,6,7,8-Tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl] benzoic acid) (TTNPB; 15 μM or 30 μM; Sigma, #T3757), 2 μg/ml oligomycin (Sigma, #O4876), carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP; 3 μM; Sigma, #C2920), 2 μM rotenone (Sigma, #R8875) and 2 μM antimycin A (Sigma, # A8674). The Seahorse protocol was as follows: Basal respiration (3 cycles; 3 min mix and 3 min measure), TTNPB injection (4 cycles; 3 min mix and 3 min measure); oligmycin (3 cycles; 3 min mix and 3 min measure); FCCP (3 cycles; 3 min mix and 3 min measure), and finally, injected together, rotenone and antimycin A (3 cycles; 3 min mix and 3 min measure). TTNPB was added as a first injection in order to activate UCP1, and then proton leak respiration was measured as that following the addition of oligomycin (to inhibit ATP synthase). To assess UCP1-dependent respiration, we compared OCR values in the presence of oligomycin from vehicle treated or TTNPB treated cells in each individual transfection condition (GFP-, UCP1wt, UCP1K−-R and UCP1K−-Q transfected HEK 293T cells).
2.11. Clark electrode based analyses of UCP-dependent respiration in BAT mitochondria
Mitochondria were re-suspended in a buffer containing (20 mM TES (pH 7.2), 100mMKCl and 0.6% fatty-acid-free BSA). Respiration rates of iBAT mitochondria were determined in Oxytherm (HansaTech, Kings Lynn, UK) Clark electrode systems at 37 °C, at 0.25 mg-0.35 mg protein/mL in100mM KCL, 20 mM TES (pH 7.2), 4 mM KH2PO4, 2 mM MgCl2, and 1 mM EDTA and 0.3% fatty acid-free bovine serum albumin. Mitochondria were energized with 25 μM palmitoyl-l-carnitine (PLC; Sigma, #P1645) or 5 mM glycerol-3-phosphate (G3P; Sigma, #G7886) plus 5 mM malate (Sigma, #M1000). Malate was added to replenish TCA cycle intermediates lost during the isolation process. After stable rates of oxygen consumption were determined, 2 mM guanosine 5′-diphosphate (GDP; Sigma, #G7127) was then added to determine UCP1-dependent respiration. Oxygen consumption rates were normalized to mitochondrial protein, as determined by Bradford assays.
2.12. High resolution respirometery of the activities of electron transport chain complexes in BAT mitochondria
Respiration rates linked to each electron transport chain complex (CI, CII, CIII and CIV), were determined using high resolution respirometry (HRR) (Oxygraph-2k; Oroboros, Innsbruck, Austria). To rule out the possibility that these respiratory rates were confounded by effects of SIRT3 on UCP1 we began each experiment by uncoupling the mitochondria by adding 1.4 μM FCCP (Sigma, #C2920) prior to the addition of substrates and inhibitors.
The Oxygraph-2k units were calibrated and all measurements were performed at 37 °C. Rates were determined for 100 μg of mitochondrial protein in 2 ml of buffer (110 mM sucrose, 60 mM K-lactobionate, 20 mM HEPES, 20 mM taurine, 10 mM KH2PO4, 3 mM MgCl2, 0.5 mM EGTA, 1 g/l BSA, pH 7.1), in the presence of 5 mM malate (Sigma, #M1000) and 1.4 μM FCCP. Rates specifically associated with each of the complexes were assessed as follows: Complex I [5 mM pyruvate (Sigma, #P2256); followed with 0.5 μM rotenone (Sigma, #R8875) to inhibit CI], Complex II [10 mM succinate (Sigma, #S2378); followed by 5 mM malonate (Sigma, #M1296) to inhibit CII], Complex III [5 mM G3P (Sigma, #G7886); followed by 1 μM antimycin A (Sigma, # A8674) to inhibit CIII], and Complex IV [0.5 mM TMPD (Sigma, # 3134) and 2 mM ascorbate (Sigma, # A4034); followed by 100 mM azide (Sigma, #S2002) to inhibit CIV]. Oxygen consumption rates (OCR) were normalized to mitochondrial protein levels, as determined by Bradford assays.
2.13. Plasma acylcarnitine analyses
To stimulate BAT thermogenesis, mice were injected with the β3-adrenergic agonist, CL316,243 (1 mg/kg; i.p.). Ninety min later, mice were anaesthetized and blood was collected by cardiac puncture for plasma acylcarnitine measurements. Blood samples were dried on filter paper (Whatman ProteinSaver 903) and stored at −20 °C. Measurements were done at the Newborn Screening lab at the Children's Hospital of Eastern Ontario (CHEO) using mass spectrometry. Methods are described in [34].
2.14. Statistical analysis
Data are represented as mean ± SEM. Two-tailed Student's t-tests, one-way ANOVA and two-way ANOVA with multi-comparison tests were used to determine statistical significance. Statistical significance, and Ns, are described in the figure legends.
2.15. Acetylation annotations
The annotation and associated analyses presented in Supplementary Table 3 were performed using the functionality of SLiMSearch tool as previously described [35]. The identified acetylation sites were annotated with overlapping sequence annotation, including information describing protein modular architecture, post-translational modifications, any solved protein structures, SNPs and experimentally characterized functional regions.
2.16. Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [36] partner repository with the dataset identifier PXD013056. The cold stress dataset is annotated under the ‘MD10’ keys and the room temperature experiment is annotated under the ‘OMDYS21-32’ keys.
3. Results
3.1. SIRT3 absence does not affect metamorphic characteristics of mice
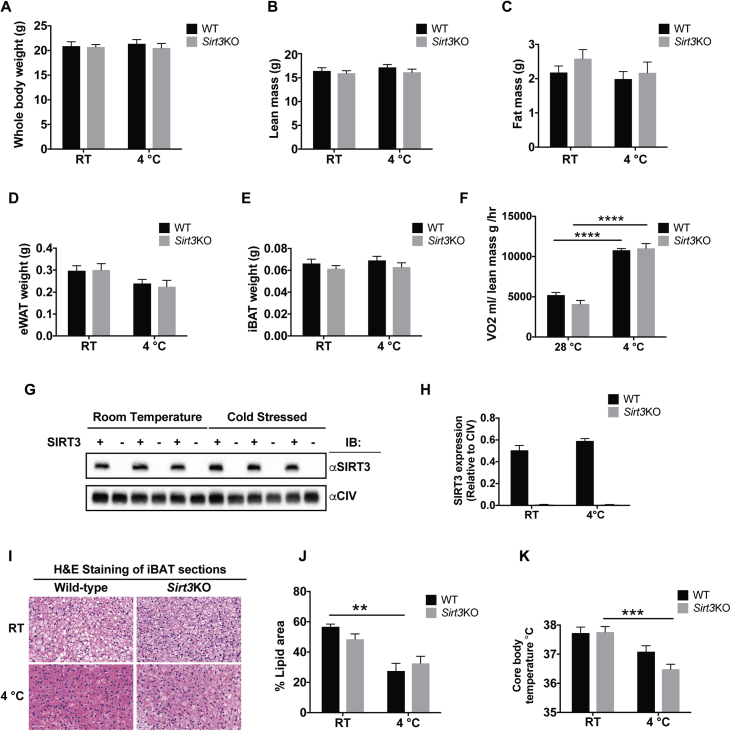
To investigate the role of SIRT3 under cold stress, we began by comparing body weight, body composition and metabolic characteristics in Sirt3KO and wild type (WT) mice at the whole body level. Previous work in mice has established that during 1 day at 4 °C, BAT is stimulated to such a high extent that shivering thermogenesis becomes minimal [37], [38], [39]. Therefore, to assess effects on BAT thermogenic processes, mice were challenged to the cold (4 °C) for 2 days. Under these conditions, we found Sirt3KO mice showed no differences in total body weight, lean mass and fat mass as determined by Echo-MRI, relative to WT mice (Figure 1A–C). Both Sirt3KO and WT mice also showed no differences in epididymal white adipose tissue (eWAT) or interscapular brown adipose tissue (iBAT) mass before and after cold exposure (Figure 1D,E). To study the effect of the cold on the resting metabolic rate, oxygen consumption (VO2) was assessed before and after cold exposure. Measurements normalized to lean body mass showed that WT and Sirt3KO mice had similarly increased metabolic rates upon cold exposure (Figure 1F). Also oxygen consumption was assessed before and after injecting mice with CL316,243. WT and Sirt3KO mice showed similar increases (Supplementary Figure 1A, B).
Figure 1.
Sirt3KO mice have normal morphometrics but impaired use of lipid in BAT and impaired thermoregulation upon cold exposure. WT and Sirt3KO mice were housed at room temperature (23 °C) or were exposed to 4 °C for 2 days. (A) Whole body weight, (B) Lean mass, (C) Fat mass, (D) Epididymal white adipose tissues (eWAT), (E) interscapular brown adipose tissues (iBAT) was measured, N = 8–9/group. (F) WT and Sirt3KO mice were kept either at 28 °C or 4 °C in CLAMS. VO2 was measured and normalized to lean body mass, N = 8–9/group. (G) Western blotting analysis of SIRT3 expression in isolated mitochondria from iBAT of either room temperature housed or cold exposed WT and Sirt3KO mice. N = 3/group. (H) Quantification of SIRT3 western blotting. (I) H & E stained sections of iBAT of room temperature housed or cold stressed WT and Sirt3KO mice. N = 4–6/group. Scale bar for all images: 50 μm. (J) Lipid droplet surface area percentage, as analyzed by quantitative morphometry. N = 4–6/group. K) Core body temperature, measured before and after cold exposure. N = 8/group. Data are represented as mean ± SEM. Two-way ANOVA with Tukey's test, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.2. SIRT3 absence results in impaired BAT lipid use and thermoregulation in mice
While there were no changes to these phenotype characteristics of Sirt3KO mice upon cold exposure at the whole body level, we also examined the metabolic and morphologic changes within BAT itself. Since it has been reported that the mRNA level of SIRT3 is increased in BAT in response to the cold [16], we first tested the impact of cold on SIRT3 protein levels by isolating mitochondria from iBAT of room temperature housed or cold exposed mice. Surprisingly, SIRT3 protein level in WT mice was not increased upon cold exposure (Figure 1G,H).
While previous work demonstrates that FAs derived from white adipose tissue (WAT) support BAT thermogenesis [40], [41], FAs are also supplied locally from BAT lipid droplets [5]. Therefore, we measured lipid droplet content in BAT of WT and Sirt3KO mice using quantitative morphometry. WT mice showed a significant reduction in lipid droplet content in iBAT sections after 2 days of cold exposure, but this same significant decrease was not observed in Sirt3KO mice (Figure 1I,J). These data suggest decreased FA utilization in the absence of SIRT3. Moreover, unlike WT mice, there was no significant decrease in the RER of Sirt3KO mice after CL316,243 injection indicating a fatty acid oxidation defect upon BAT activation (Supplementary Figure 1C, D). Consistent with a functional impact on lipid use and fatty acid oxidation when BAT is activated, Sirt3KO mice had a significant reduction in their ability to maintain core body temperature in response to the cold (Figure 1K). Therefore, while Sirt3KO mice were outwardly phenotypically normal, they had defects consistent with a role for SIRT3 in thermoregulation. Next, we investigated the functional relationship between SIRT3 and BAT mitochondrial respiration.
3.3. Impaired respiration in BAT mitochondria of Sirt3KO mice
To study UCP1-dependent proton leak respiration, isolated iBAT mitochondria from room temperature housed WT and Sirt3KO mice were energized by 25 μM palmitoyl-l-carnitine (PLC) since long chain fatty acids are activators and substrates for BAT mitochondria [5], [42]. UCP1 activity was subsequently inhibited by adding 2 mM guanosine diphosphate (GDP). Mitochondria isolated from BAT of Sirt3KO mice showed defective UCP1-dependent respiration relative to those from WT mice (Figure 2A). Moreover, Sirt3KO mitochondria function was examined using glycerol-3-phosphate (G3P), which is an excellent substrate for BAT mitochondrial respiration [43] and provides reducing equivalents that enter the electron transport chain at Complex III. BAT mitochondria from either room temperature housed or cold exposed WT and Sirt3KO mice were energized by 5 mM G3P and UCP1 activity was then inhibited by adding 2 mM GDP. Under these conditions, UCP1-dependent respiration in Sirt3KO mitochondria was lower than that in WT mice (Figure 2B,C). UCP1 levels after cold exposure were increased by cold stress in both WT and Sirt3KO mice, but we found no observable difference between genotypes (Figure 2D,E). Thus, the absence of SIRT3 itself did not affect the expression of UCP1 protein, suggesting that the decrease in UCP1-dependent respiration in Sirt3KO mice might be due to changes in UCP1 activity.
3.4. Cold stress and SIRT3 induce acetylation and deacetylation, respectively, of BAT mitochondrial proteins
We reasoned that SIRT3 could impact UCP1 activity directly by regulating acetylation of UCP1 lysine residues, or indirectly by regulating critical pathways required for UCP1 function (e.g., upstream oxidation reactions that supply electrons to the respiratory chain, or the respiratory chain itself). To investigate these potential modes of regulation, we used label-free quantitative acetylome profiling to compare non-histone protein acetylation in iBAT mitochondria isolated from WT or Sirt3KO mice (Figure 3A). SIRT3 impacted the acetylation status of hundreds of sites (or site combinations) in separate experiments carried out for room temperature housed or cold stressed mice (Supplementary Table 1). There was a significant correlation between acetylome profiles in room temperature and cold stressed mice (Figure 3B & Supplementary Figure 2A). As expected based on the role of SIRT3 as a lysine deacetylase, the vast majority of impacted acetylation marks were upregulated rather than downregulated in Sirt3KO mice (Figure 3B & Supplementary Figure 2A). This was confirmed by western blotting experiments (Figure 3C,D). For most of the significantly upregulated acetylations, the observed fold change (log2FC Sirt3KO/WT) seemed to be more dramatic at room temperature than in the cold (Figure 3E & Supplementary Figure 2B). This may be explained in part by the intriguing observation that cold stress itself increases the baseline of acetylation in wild-type mice (Figure 3C,D). Since our room-temperature and cold-stressed samples were processed for mass spectrometry at different times, identification of those acetylations impacted by cold-stress independently of SIRT3 will require additional experiments. Notably, analysis of non-acetylated peptides recovered in our experiments suggested that changes in acetylation status could not be explained by changes in protein levels (Supplementary Figure 3A, B & Supplementary Table S2).
GO-term enrichment of statistically significant upregulated sites (log2FC Sirt3KO/WT = ≥ 1, adjusted P value ≤ 0.05) using Metascape indicated enrichment of proteins assigned to fatty acid metabolic processes and the mitochondrial matrix under room temperature and cold stressed conditions (Figure 3F & Supplementary Figure 2C). Cold stressed mice also showed preferential SIRT3-regulation of proteins functioning in the tricarboxylic acid (TCA) cycle (Figure 3F & Supplementary Figure 2C). Altogether, our data point to a significant contribution for SIRT3 in the regulation of the BAT mitochondrial acetylome, with enrichment in pathways that are likely to intersect with UCP1 function. Functional annotation of acetylation sites uncovered in our work using the SLiMSearch tool uncovered many sites in close proximity to known regulatory regions, domains, motifs and other previously identified PTMs (Supplementary Table S3).
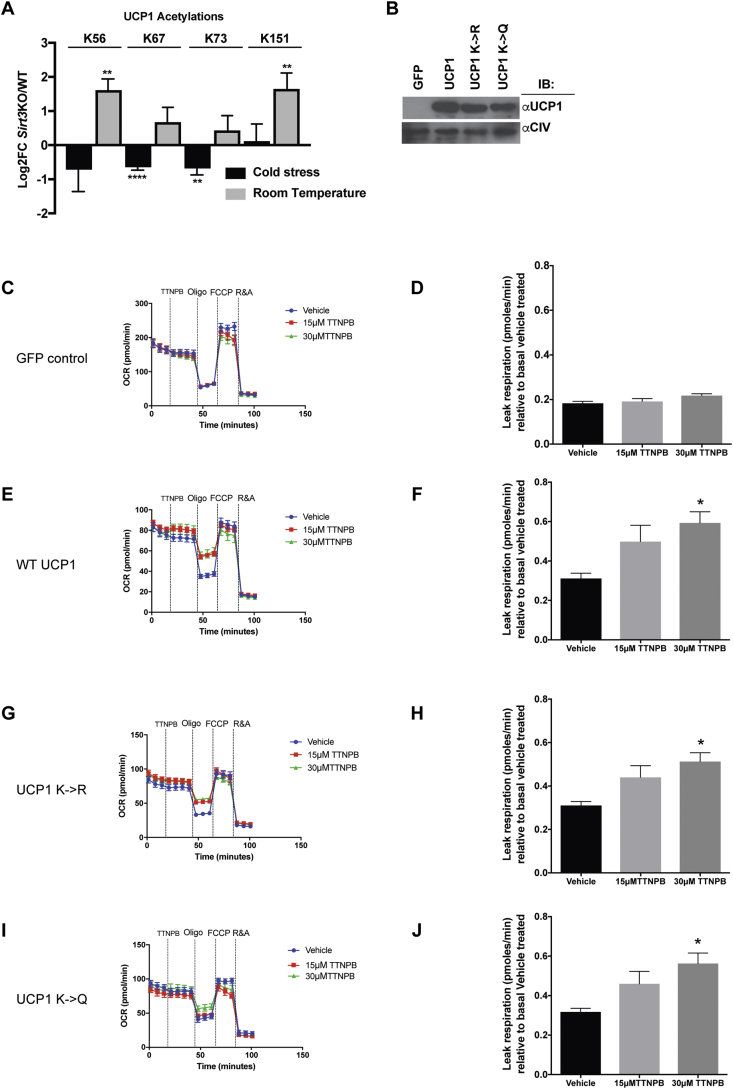
3.5. Acetylation of lysines on UCP1 does not affect UCP1 leak respiration
In support of acetylation playing a direct role in UCP1 function within BAT mitochondria, we uncovered four acetylations on UCP1 in both room temperature and cold stressed mice (K56, K67, K73 and K151). K56 and K151 showed a significant increase in acetylation in samples from Sirt3KO mice, but only when housed at room temperature (∼3-fold each, Figure 4A & Supplementary Table 1). Intriguingly, these residues are located in similar positions in the first two of the three matrix-side loops of the protein [44], and theoretically would be accessible to matrix SIRT3 activity. To determine if acetylation of K56 and K151 are important for UCP1-dependent leak respiration, we established an in vitro assay expressing mouse UCP1 in HEK293T cells. UCP1 function has previously been examined in the HEK293 cell system [45], [46], [47]. Cells were transfected with plasmids expressing wild-type murine UCP1(WT) or mutant UCP1 variants in which these residues cannot be acetylated, i.e., UCP1 (K→ R) or that mimic constitutive acetylation, i.e., UCP1 (K→ Q). A plasmid expressing GFP from the same vector was used as a negative control. Importantly, since HEK293T cells do not express UCP1 endogenously, these constructs provided the only source of UCP1, and the UCP1 variants were expressed at similar levels (Figure 4B). Herein, we used the retinoic acid analogue, TTNPB, to activate UCP1 in intact HEK293T cells rather than long chain FAs. In brown adipocytes, unlike HEK293 cells, long chain FAs are rapidly taken up and then used for the activation of UCP1, and fueling of beta-oxidation. Previous work has demonstrated that the fatty acid transporter, CD36, is not expressed in HEK293 cell [48] as it is in brown adipocytes. Thus we used TTNPB, which is cell permeable, to induce UCP1 leak respiration, as used before in UCP1 expressing HEK293 cells in plate-based respirometry assays [47], [49], [50].
Figure 4.
Acetylation of lysines on UCP1 does not affect UCP1 leak respiration. (A) Regulated sites were detected in UCP1 from BAT mitochondria isolated from room temperature housed WT and Sirt3KO mice. Error bars represent standard error of the mean, and corrected p values were determined by MSstats, ** p <0.01, **** p < 0.0001. In corresponding cold stressed WT and Sirt3KO mice, no sites with statistically-significant changes were detected. (B) Western blotting of UCP1 expression in HEK293T cells that were transfected with GPF, UCP1(wt), UCP1(K- > R) and UCP1 (K- > Q) plasmids. C) A representative trace of functional analyses of GFP transfected cells that were injected with a vehicle control or injected with 15 μM or 30 μM TTNPB. D) UCP1 leak respiration measurements of GFP transfected cells. N = 4. E) A representative trace of functional analyses of UCP1(wt) transfected cells that were injected with a vehicle control or injected with 15 μM or 30 μM TTNPB. F) Leak respiration measurements of UCP1(wt) transfected cells. N = 4. G) A representative trace of functional analyses of UCP1(K- > R) transfected cells that were injected with a vehicle control or injected with 15 μM or 30 μM TTNPB. H) UCP1 leak respiration measurements of UCP1(K- > R) transfected cells. N = 4. I) A representative trace of functional analyses of UCP1 (K- > Q) transfected cells that were injected with a vehicle control or injected with 15 μM or 30 μM TTNPB. J) UCP1 leak respiration measurements of UCP1 (K- > Q) transfected cells. N = 4. All the trace graphs are representative of four independent experiments and UCP1 leak graphs are an average of the four independent experiments. Data are represented as mean ± SEM. One-way ANOVA with Bonferroni's test was used; *p < 0.05.
As expected, GFP transfected cells did not show TTNPB-induced respiration (Figure 4C,D). In contrast, UCP1(WT) transfected cells had increased UCP1-mediated leak respiration (Figure 4E,F). A similar effect on leak respiration was observed with acetyl-defective UCP1 (Figure 4G,H) and with acetyl-mimic forms of UCP1 (Figure 4I,J), suggesting that these lysines alone cannot account for the impact of SIRT3 on UCP1 dependent respiration. Therefore, we investigated if the lower rate of UCP1-dependent respiration in Sirt3KO mitochondria was instead due to SIRT3-mediated regulation of upstream proteins of UCP1 that are involved in vital pathways supporting UCP1-mediated thermogenesis. Specifically, we focused on the FAO and ETC pathways.
3.6. Decreased plasma levels of medium and long chain acylcarnitines in Sirt3KO mice
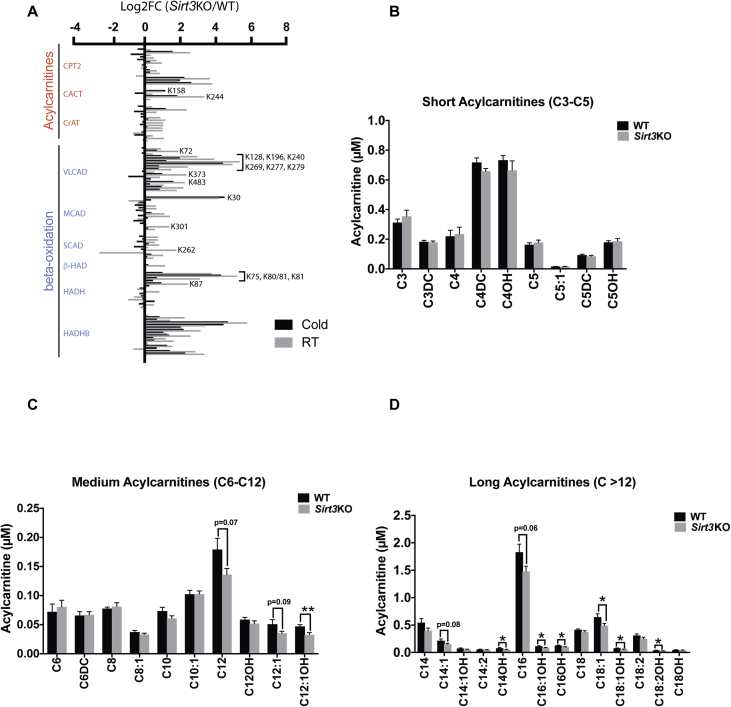
Beyond being an allosteric agonist of UCP1 activity, FAs are a major energy substrate for active BAT. To be taken up into mitochondrial for oxidation, long chain FAs are first converted to acylcarnitines and then are transported to mitochondria matrix via carnitine acylcarnitine translocase (CACT) [51]. Defective FAO can lead to increased circulating levels of acylcarnitines, and fasted Sirt3KO mice have previously been shown to have a defect in liver mitochondrial FAO and an accumulation of long chain acylcarnitines in the plasma [21]. In our study, our acetylome profiling data showed that SIRT3 negatively regulated lysine acetylations on many proteins involved in the uptake and oxidation of fatty acids and in the metabolism of acylcarnitines in BAT from both room temperature housed and cold exposed mice (Figure 5A & Supplementary Table 1). Many of the lysine residues with strongly-regulated acetylations are located in important functional regions of target proteins, as identified by our SlimSearch annotation (Figure 5A and Supplementary Table S3). Regulation of these acetylations may be particularly important for FAO and acetylcarnitine metabolism.
Figure 5.
Sirt3KO mice have decreased levels of medium- and long-chain acylcarnitines. A) Measured Log2FCs of acetylation sites in selected proteins related to fatty acid oxidation or acylcarnitine metabolism from iBAT mitochondria isolated from room temperature or cold stressed WT and Sirt3KO mice. N = 6/group. Indicated sites are those located in “regions of interest” from supplementary table 3 with ≥2 increase in Sirt3KO mice. Only data with measured values for both RT and cold stress are shown. B–D) Plasma acylcarnitine analyses of WT and Sirt3KO mice that were injected with CL316.243 (1 mg/kg i.p.) to acutely activate BAT. Cardiac puncture blood samples were collected 90min-post injection used for MS/MS based acylcarnitines analysis. B) Measurement of short-chain (C3-C5), C) medium-chain (C6-C12), D) long-chain acylcarnitines (C > 12). N = 8/group. Data are represented as mean ± SEM. Student's t-test; Two-tailed; *p < 0.05, **p < 0.01.
To examine the impact of SIRT3 absence on the level or the production of acylcarnitines when BAT is activated, mice were injected with a standard bolus of the β3-adrenergic receptor selective agonist CL 316,243 (1 mg/kg i.p.), to induce BAT activity. Blood samples were collected 90 min post-injection by cardiac puncture for MS/MS acylcarnitine analyses. These experiments revealed that Sirt3KO mice had no differences in short chain acylcarnitines (Figure 5B), but had decreased levels of select medium-chain (C12:1OH) and long-chain (C14OH, C16:1OH, C16OH, C18:1, C18:1OH, C18:2OH) acylcarnitines, compared to WT mice (Figure 5C,D).
3.7. Impaired activities of ETC protein complexes in Sirt3KO BAT mitochondria
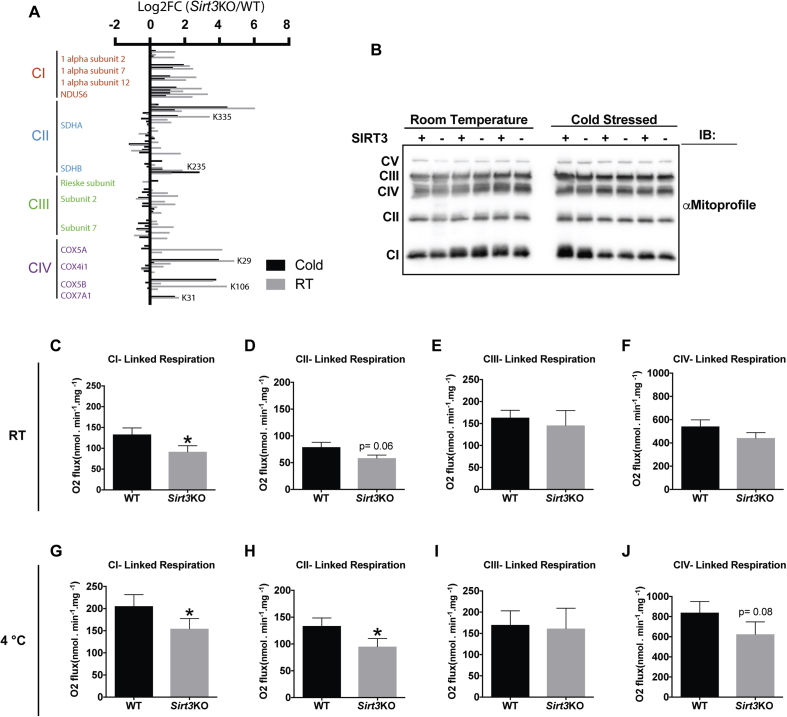
ETC proteins have a fundamental role in UCP1-dependent respiration because their activities drive the production of protonmotive force, which is necessary for UCP1 activity. Our acetylome profiling showed that proteins from each of the ETC complexes have many SIRT3 regulated sites (Figure 6A). Many of these were located in defined domains, motifs or other regions of interest (Figure 6A and Supplementary Table 3). Western blotting analysis showed that SIRT3 absence does not affect the expression levels of marker proteins for each of the ETC complexes between WT and Sirt3KO mice (Figure 6B & Supplementary Figure 4A-D). However, there was an effect of cold on the expression of the CIII marker protein, which reached statistical significance in WT mice, but not in Sirt3KO. To determine if there were functional differences in the activities of the complexes (CI, CII, CIII and CIV), we designed UCP1-independent respiration protocols in which we were able to examine each complex individually. Analyses of CI, CII, CIII and CIV activities were conducted using high resolution respirometry (O2K-Oroboros) in isolated mitochondria from BAT of WT or Sirt3KO room temperature housed (Figure 6C–F) and cold exposed mice (Figure 6G–J). The chemical uncoupler carbonylcyanide p-triflouromethoxyphenylhydrazone (FCCP), was added at the beginning of each respiration experiment to exclude any involvement of UCP1 function in the analyses. Relative to WT controls, CI was impaired in BAT mitochondria of Sirt3KO mice in room temperature [31% decreased versus WT; (Figure 6C)] and both CI and CII were decreased under cold stressed conditions [25% and 29%, respectively; (Figure 6G,H)]. Taken together, these results indicate that the absence of SIRT3 results in decreased activities of complexes I and II in BAT.
Figure 6.
Sirt3KO mice have impaired ETC function. (A) Measured Log2FCs of lysine acetylation sites in selected proteins from ETC complexes CI-CIV from isolated mitochondria from iBAT of room temperature housed or cold exposed WT or Sirt3KO mice. N = 6/group. Indicated sites are those located in “regions of interest” from supplementary table 3 with ≥2 increase in Sirt3KO mice. Only data with measured values for both RT and cold stress are shown. (B) Western blotting analysis of OXPHOS protein expression in isolated BAT mitochondria. N = 3/group. (C–F) High resolution respirometry functional analyses of ETC proteins in mitochondria isolated from BAT of room temperature housed WT and Sirt3KO mice. (C) Complex I (CI) linked respiration; (D) Complex II (CII) linked respiration; (E) Complex III (CIII) linked respiration; (F) Complex IV (CIV) linked respiration. N = 6–14. (G–J) High resolution respirometry functional analyses of ETC proteins in mitochondria isolated from BAT of cold exposed WT and Sirt3KO mice. (G) Complex I (CI) linked respiration; (H) Complex II (CII) linked respiration; (I) Complex III (CIII) linked respiration; (J) Complex IV (CIV) linked respiration. N = 6–12. Data are presented as mean ± SEM. Student's t-test; Two-tailed; *p < 0.05.
4. Discussion
4.1. Deacetylation of upstream proteins of UCP1 as a regulator of BAT thermogenesis
The control of BAT thermogenesis can be exerted at many levels: total amount of tissue; number of brown adipocytes in each depot; number of mitochondria in each cell; levels of UCP1 protein per mitochondrion; and finally the degree to which UCP1 is activated acutely by effectors such as FA upon adrenergic activation [5], [52]. Work from the Spiegelman lab also provides evidence that UCP1 mediated thermogenesis is regulated by reactive oxygen spices (ROS) and cysteine sulfenylation of UCP1 [53]. Recently, it has been reported that BAT thermogenesis can be regulated independently of adrenergic activation by unexpected metabolites such as succinate, and its oxidation by CII activates ROS induced thermogenesis [54]. However, an unanswered question is whether deacetylation of UCP1 and/or the upstream pathways of UCP1, i.e., the supply and flux of electrons from FA oxidative reactions, controls BAT thermogenesis.
Our work is the first to identify and elaborate the connections between SIRT3, the master deacetylase in the mitochondria, and BAT thermogenesis in mice. We provide evidence that the absence of SIRT3: (1) impairs use of BAT lipid content, (2) impairs thermoregulation in mice (3) increases BAT mitochondrial protein acetylation, (3) increases UCP1 lysine acetylation, but the mutation of these sites does not affect UCP1 leak respiration, (4) increases lysine acetylation in proteins that play key roles in FA uptake and oxidation, and that this is associated with decreases in circulating medium- and long-acylcarnitines, and finally, (5) increases the lysine acetylation of ETC complex subunits, which is associated with their impaired activities. Thus, we provide strong evidence that SIRT3-mediated deacetylation is fundamentally important in driving BAT thermogenesis and SIRT3 is indirectly regulating UCP1 thermogenesis.
Previous work demonstrates that that non-enzymatic mitochondrial acetylation is driven by high levels of acetyl-CoA produced by FAO [25]. It has also been suggested that SIRT3 may play a critical role in promoting the reversal of non-enzymatic acetylations [26]. Altogether, our work is consistent with a model wherein a rise in acetyl CoA levels driven by cold stress results in increased non-enzymatic acetylations. Here, we propose that UCP1-mediated thermogenesis is compromised due to a failure to deacetylate substrate uptake and oxidation proteins that function upstream of UCP1 in Sirt3KO mice. This model does not lessen the centrality of UCP1 activation in the control of BAT thermogenesis, but demonstrates the importance of fatty acid uptake and oxidation reactions, and of subsequent redox reactions in the ETC, which collectively generate the heat when BAT is activated.
4.2. Phenotypes of Sirt3KO mice
While we ultimately defined SIRT3 as a critical regulator of BAT thermogenesis, we observed no differences in body composition, elevation of VO2 upon cold exposure or CL316,243 injection, iBAT weight, mitochondrial content and UCP1 content between WT and Sirt3KO mice. Our results are supportive and consistent with previous findings that Sirt3KO mice show normal body weight [15]. However, Sirt3KO mice have impaired BAT lipid use upon cold exposure and do not show a significant decrease RER after BAT-specific activation as seen in WT mice, which could be an indicator of less activated BAT in Sirt3KO mice. Hirschey et al. previously showed that acute cold stress (6 h) is associated with cold intolerance in Sirt3KO mice, but only when mice were fasted [21]. Here we have demonstrated that Sirt3KO mice under normal housing conditions with ad libitum access to food are cold-intolerant at 48h, a time at which BAT thermogenesis is normally the major thermoregulatory mechanism and shivering thermogenesis is normally minimal. Interestingly, VO2 is not different between WT and Sirt3KO mice. This was unexpected, but given that these analyses were conducted in a whole body knockout mice, it was not entirely surprising since metabolic responses to cold are highly complex and represent an aggregated consequence of the effects of cold on many systems (e.g., neural, muscular, circulatory). It is possible that the thermoregulatory phenotype of the whole body Sirt3KO mice is affected by the absence of SIRT3 in tissues other than BAT. In spite of the complexity of the study model, specific activation of BAT by a β3-adrenergic receptor agonist proved that the activity of BAT in Sirt3KO mice is impaired. It is also possible that Sirt3KOmice have defective regulation of heat loss as seen in other mouse models in which altered heat loss contributes to thermoregulation in mice [55].
4.3. Mechanism of SIRT3 function in BAT thermogenesis
UCP1 is essential for BAT thermogenesis, and induction of UCP1 in Sirt3KO mice was similar to that observed in WT mice. This observation contrasts with previous work in vitro. Specifically, Shi et al. reported that overexpression of SIRT3 in the brown adipocyte cell line, HIB1B, resulted in increased expression of UCP1 mRNAs and that overexpression of dominant negative forms of SIRT3 leads to down-regulated expression of UCP1 [16]. Instead, our observation is consistent with the in vivo findings of Lombard and colleagues who showed that the absence of SIRT3 does not affect the expression of UCP1 in mouse BAT [15]. Despite the wild-type expression level of UCP1, UCP1-mediated respiration in mitochondria from Sirt3KO mice was significantly decreased under cold stress. This was not observed in room temperature housed animals. Cold stress may provide an optimal scenario under which to evaluate the function of UCP1 when it is fully activated, and it was in these conditions that we found defective UCP1 mediated respiration in Sirt3KO mice. Our acetylome work suggests that the functional relationship between UCP1 and SIRT3 stems from an impact of SIRT3 on upstream regulatory pathways.
4.4. Regulated acetylations driving thermogenesis
We identified four acetylation sites on UCP1 in mitochondria from mouse BAT (K56, K67, K73 and K151) and this is consistent with a previous study that identified three acetylation sites (K67, K73 and K151) in UCP1 in BAT homogenate from rat [56]. Mutation of the two SIRT3-regulated sites (K56 and K151) did not impact UCP1 function in a cell culture system. Yet, we cannot completely rule out a role for acetylation in the direct regulation of UCP1. Point mutants of UCP1 were assayed in a HEK293T cell line, which has previously been used to study the effects of novel activators of UCP1, such as TTNPB [47]. However, this system may not account for variables that normally play important roles in UCP1 function in brown adipocytes. For example, the HEK293T cell model clearly cannot fully recapitulate the effects of cold on signaling and metabolic processes that occur in vivo. We also focused exclusively on acetylation sites that face the mitochondrial matrix. Other acetylations that were not tested or that were not identified in our work could also be important for UCP1 function. Finally, UCP1 acetylation/deacetylation could function redundantly with other PTMs such as sulfenylation [53], or indirect modes of UCP1 regulation that we describe here. Notably, our observation that SIRT3 did not impact UCP1 acetylation sites in cold stress suggests that any such regulation is unlikely to account for the impact of SIRT3 on UCP1-dependent respiration.
BAT oxidizes a large amount of FAs from the circulation originating from white adipose tissue (WAT) or from the lipolysis of lipid droplets in BAT. Defects in FAO cause the release of acylcarnitines from mitochondria into the circulation [57], [58], and given our mitochondrial acetylome results, we originally hypothesized that circulating acylcarnitines would be increased. However, our results show that the plasma levels of medium-chain and long-chain acylcarnitines in Sirt3KO mice were lower compared to WT mice. The following interpretations are possible: 1) decreased formation of acylcarnitines, which requires the uptake of FA by BAT mitochondria, and/or, 2) increased consumption of acylcarnitines by BAT mitochondria. We favor the former, given that our results show the acetylation of many of the enzymes involved in acylcarnitine/fatty acid metabolism in BAT mitochondria, which could result in a negative impact on enzymatic activity. For example, our acetylome profiling revealed that CACT, the main mitochondrial enzyme that regulates the entry of acylcarnitines to the mitochondria for FAO processes, was hyper-acetylated in Sirt3KO mitochondria and this could decrease its function. However, other interpretations are possible: there may be indirect effects of the activation of BAT thermogenesis by the use of CL compound, wherein increases in circulating free fatty acids from WAT or BAT result in genotype-specific differences in acylcarnitine metabolism in other tissues, such as the liver. Further research is needed to clarify the role of SIRT3 in acylcarnitine metabolism in activated BAT.
Multi-tissue quantitative acetylome work shows that SIRT3 expression varies between tissues such as liver, heart, kidney, brain, and skeletal muscle [59]. Moreover, the mitochondrial regulation mediated by SIRT3 in various tissues is in a tissue-specific fashion and there are differences in the degree of how much SIRT3 regulates the metabolic pathways in different tissues [59], suggesting the importance of studying the role of SIRT3 in regulating the metabolic pathways in BAT and in its activated status. Given the unique function and important physiological role of BAT, we studied the role of SIRT3 in regulating the ETC pathway in BAT mitochondria, which is not well-characterized. For the first time, we report that Sirt3KO mitochondria from iBAT of cold exposed mice have impaired activities of CI and CII during cold stress and this correlates with the increased acetylation observed on subunits from CI and CII proteins. These observations extend the work of other groups who show that CI and/or CII function are decreased in isolated mitochondria from liver of Sirt3KO mice [60], [61]. Finley et al. found a trend (p = 0.07) for decreased enzymatic activity of CII in BAT mitochondria from Sirt3KO mice, but these analyses were not carried out under conditions that activate BAT, and more importantly, they were spectrophotometric Vmax-type assays, rather than assays of its activity within the functioning respiratory chain [62]. Although previous work identified SIRT3 regulated sites in Subunit A of CII in liver mitochondria [61], [62], here our acetylome profiling data identify SIRT3 regulated lysine acetylation sites in subunit A and subunit B of CII in BAT mitochondria from mice housed in room temperature and cold conditions, suggesting a mechanism for the impact of SIRT3 on CII function specifically in this tissue. While we show that CI and CII proteins are equally expressed in BAT mitochondria from WT and Sirt3KO mice, we cannot rule out the possibility that the functional defects described herein stem from decreased stability of some subunits of CI and CII that were not examined. Alternatively, hyper-acetylation in the absence of SIRT3 may impair complex functions without affecting overall protein levels. An unresolved question is the fraction of each target protein population that is acetylated in the presence and absence of SIRT3. Previous efforts to quantify stoichiometry of individual acetylation sites suggests that even regulated acetylations often involve only a small fraction of the target population at any one time [26], [63], [64]. In the mitochondria, these low-level acetylations may represent a form of protein damage that disrupt critical functions only when a threshold is reached across many proteins in a given pathway [26]. On the other hand, acetylations impacting a larger fraction of a target population may be better situated to disrupt critical protein–protein interactions or directly inhibit important enzymatic activities.
5. Conclusion
Altogether, our results demonstrate for the first time that SIRT3 mediated deacetylation of an extensive array of mitochondrial proteins in BAT plays a fundamental role in thermogenesis. We propose a model wherein UCP1-mediated thermogenesis is indirectly regulated by deacetylation of many substrate uptake and oxidation pathways that are upstream of UCP1. Our proteomics findings provide an extensive resource for future investigations into these mechanisms, and others, in the context of the newly described functional connections between SIRT3 and UCP1.
Author contributions
RS, MD and MEH conceived the ideas for the project. RS conducted most of the experiments and analyzed most of the results. MD helped with mass spectrometry (MS) sample preparation, analyzed MS data and prepared MS related figures. JJ conducted mass spectrometry analysis. CP helped with many high resolution respirometry analyses of the electron transport chain complexes. MN helped with western blotting and histological analyses. JX helped with in vivo analyses of mice, genotyping and colony maintenance. YS helped with MS sample prep. EBC helped to design UCP1 constructs and contributed expertise for cell culture. QT provided Sirt3KO mice and expert advice. NK supervised the work of JJ. IK and NED carried out the acetylation annotation analysis. RS, MD and MEH drafted and wrote the paper. All authors read and approved the paper.
Acknowledgements
Authors are very grateful for the expert advice of Dr. Martin Jastroch (Wenner-Gren Institute, Stockholm University) during the optimization of Seahorse analyses of UCP1 transfected HEK293T cells. We would like to thank the Newborn Screening group at the Children's Hospital of Eastern Ontario for their expert assistance with acylcarnitine analyses. We thank Alix Denoncourt for help with plasmid prep. We thank Ziyad El Hankouri for help with western blotting. This work was funded by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to MEH; RGPIN 04973); NSERC grant RGPIN 05015 (to MD), NIH grants: U19AI118610 and P01CA177322 to JRJ and P50 GM082250 to NK. RS is the recipient of a scholarship from the University of Shaqra, Duwadimi, Saudi Arabia. MN was funded by an NSERC Undergraduate Research Scholarship. CP is the recipient of a Mitacs postdoctoral fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.04.008.
Contributor Information
Michael Downey, Email: mdowne2@uottawa.ca.
Mary-Ellen Harper, Email: mharper@uottawa.ca.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Blondin D.P., Labbé S.M., Tingelstad H.C., Noll C., Kunach M., Phoenix S. Increased brown adipose tissue oxidative capacity in cold-acclimated humans. Journal of Clinical Endocrinology & Metabolism. 2014;99(3):E438–E446. doi: 10.1210/jc.2013-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Himms-Hagen J. Brown adipose tissue metabolism and thermogenesis. Annual Review of Nutrition. 1985;5(1):69–94. doi: 10.1146/annurev.nu.05.070185.000441. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls D.G., Locke R.M. Thermogenic mechanisms in brown fat. Physiological Reviews. 1984;64(1):1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 4.U Din M., Raiko J., Saari T., Kudomi N., Tolvanen T., Oikonen V. Human brown adipose tissue [(15)O]O2 PET imaging in the presence and absence of cold stimulus. European Journal of Nuclear Medicine and Molecular Imaging. 2016;43(10):1878–1886. doi: 10.1007/s00259-016-3364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 6.Bartelt A., Bruns O.T., Reimer R., Hohenberg H., Ittrich H., Peldschus K. Brown adipose tissue activity controls triglyceride clearance. Nature Medicine. 2011;17(2):200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 7.Yu X.X., Lewin D.A., Forrest W., Adams S.H. Cold elicits the simultaneous induction of fatty acid synthesis and β-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. The FASEB Journal. 2002;16(2):155–168. doi: 10.1096/fj.01-0568com. [DOI] [PubMed] [Google Scholar]
- 8.Enerbäck S., Jacobsson A., Simpson E.M., Guerra C., Yamashita H., Harper M.-E. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387(6628):90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 9.Heaton G.M., Wagenvoord R.J., Kemp A., Nicholls D.G. Brown-Adipose-Tissue mitochondria: photoaffinity labelling of the regulatory site of energy dissipation. European Journal of Biochemistry. 1978;82(2):515–521. doi: 10.1111/j.1432-1033.1978.tb12045.x. [DOI] [PubMed] [Google Scholar]
- 10.Rabelo R., Schifman A., Rubio A., Sheng X., Silva J.E. Delineation of thyroid hormone-responsive sequences within a critical enhancer in the rat uncoupling protein gene. Endocrinology. 1995;136(3):1003–1013. doi: 10.1210/endo.136.3.7867554. [DOI] [PubMed] [Google Scholar]
- 11.Shore A., Emes R.D., Wessely F., Kemp P., Cillo C., D'Armiento M. A comparative approach to understanding tissue-specific expression of uncoupling protein 1 expression in adipose tissue. Frontiers in Genetics. 2013;3 doi: 10.3389/fgene.2012.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao W., Daniel K.W., Robidoux J., Puigserver P., Medvedev A.V., Bai X. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the Brown fat uncoupling protein 1 gene. Molecular and Cellular Biology. 2004;24(7):3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholls D.G., Rial E. A history of the first uncoupling protein, UCP1. Journal of Bioenergetics and Biomembranes. 1999;31(5):399–406. doi: 10.1023/a:1005436121005. [DOI] [PubMed] [Google Scholar]
- 14.Shabalina I.G., Jacobsson A., Cannon B., Nedergaard J. Native UCP1 displays simple competitive kinetics between the regulators purine nucleotides and fatty acids. Journal of Biological Chemistry. 2004;279(37):38236–38248. doi: 10.1074/jbc.M402375200. [DOI] [PubMed] [Google Scholar]
- 15.Lombard D.B., Alt F.W., Cheng H.-L., Bunkenborg J., Streeper R.S., Mostoslavsky R. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and Cellular Biology. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi T., Wang F., Stieren E., Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in Brown adipocytes. Journal of Biological Chemistry. 2005;280(14):13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 17.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H. Sirt5 is an NAD-dependent protein lysine demalonylase and desuccinylase. Science (New York, N.Y.) 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haigis M.C., Mostoslavsky R., Haigis K.M., Fahie K., Christodoulou D.C., Murphy A.J. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 19.Tan M., Peng C., Anderson K.A., Chhoy P., Xie Z., Dai L. Lysine glutarylation is a protein post-translational modification regulated by SIRT5. Cell Metabolism. 2014;19(4):605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson K.A., Hirschey M.D. Mitochondrial protein acetylation regulates metabolism. Essays in Biochemistry. 2012;52 doi: 10.1042/bse0520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B. SIRT3 regulates fatty acid oxidation via reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirschey M.D., Shimazu T., Huang J.-Y., Schwer B., Verdin E. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harbor Symposia on Quantitative Biology. 2011;76:267–277. doi: 10.1101/sqb.2011.76.010850. [DOI] [PubMed] [Google Scholar]
- 23.Jing E., O'Neill B.T., Rardin M.J., Kleinridders A., Ilkeyeva O.R., Ussar S. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62(10):3404–3417. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinert B.T., Iesmantavicius V., Moustafa T., Schölz C., Wagner S.A., Magnes C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Molecular Systems Biology. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pougovkina O., te Brinke H., Ofman R., Cruchten V.,G.,A., Kulik W., Wanders R.J. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Human Molecular Genetics. 2014;23(13):3513–3522. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- 26.Weinert B.T., Moustafa T., Iesmantavicius V., Zechner R., Choudhary C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. The EMBO Journal. 2015;34(21):2620–2632. doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palacios O.M., Carmona J.J., Michan S., Chen K.Y., Manabe Y., Ward J.L., III Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1α in skeletal muscle. Aging (Albany NY) 2009;1(9):771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shabalina I.G., Ost M., Petrovic N., Vrbacky M., Nedergaard J., Cannon B. Uncoupling protein-1 is not leaky. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797(6):773–784. doi: 10.1016/j.bbabio.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Downey M., Johnson J.R., Davey N.E., Newton B.W., Johnson T.L., Galaang S. Acetylome profiling reveals overlap in the regulation of diverse processes by sirtuins, gcn5, and esa1. Molecular & Cellular Proteomics: MCP. 2015;14(1):162–176. doi: 10.1074/mcp.M114.043141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 32.Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics (Oxford, England) 2014;30(17):2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- 33.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host & Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beauchamp B., Thrush A.B., Quizi J., Antoun G., McIntosh N., Al-Dirbashi O.Y. Undernutrition during pregnancy in mice leads to dysfunctional cardiac muscle respiration in adult offspring. Bioscience Reports. 2015;35(3) doi: 10.1042/BSR20150007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krystkowiak I., Davey N.E. SLiMSearch: a framework for proteome-wide discovery and annotation of functional modules in intrinsically disordered regions. Nucleic Acids Research. 2017;45 doi: 10.1093/nar/gkx238. Web Server issue): W464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golozoubova V., Hohtola E., Matthias A., Jacobsson A., Cannon B., Nedergaard J. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. The FASEB Journal. 2001;15(11):2048–2050. doi: 10.1096/fj.00-0536fje. [DOI] [PubMed] [Google Scholar]
- 38.Nedergaard J., Cannon B. UCP1 mRNA does not produce heat. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2013;1831(5):943–949. doi: 10.1016/j.bbalip.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Stier A., Bize P., Habold C., Bouillaud F., Massemin S., Criscuolo F. Mitochondrial uncoupling prevents cold-induced oxidative stress: a case study using UCP1 knockout mice. Journal of Experimental Biology. 2014;217(4):624–630. doi: 10.1242/jeb.092700. [DOI] [PubMed] [Google Scholar]
- 40.Schreiber R., Diwoky C., Schoiswohl G., Feiler U., Wongsiriroj N., Abdellatif M. Cold-induced thermogenesis depends on ATGL-mediated lipolysis in cardiac muscle, but not Brown adipose tissue. Cell Metabolism. 2017;26(5):753–763. doi: 10.1016/j.cmet.2017.09.004. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin H., Ma Y., Chanturiya T., Cao Q., Wang Y., Kadegowda A.K.G. Lipolysis in Brown adipocytes is not essential for cold-induced thermogenesis in mice. Cell Metabolism. 2017;26(5):764–777. doi: 10.1016/j.cmet.2017.09.002. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva J.P., Shabalina I.G., Dufour E., Petrovic N., Backlund E.C., Hultenby K. SOD2 overexpression: enhanced mitochondrial tolerance but absence of effect on UCP activity. The EMBO Journal. 2005;24(23):4061–4070. doi: 10.1038/sj.emboj.7600866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shabalina I.G., Petrovic N., de Jong J.M.A., Kalinovich A.V., Cannon B., Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Reports. 2013;5(5):1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 44.Klingenberg M., Echtay K.S., Bienengraeber M., Winkler E., Huang S.G. Structure-function relationship in UCP1. International Journal of Obesity and Related Metabolic Disorders: Journal of the International Association for the Study of Obesity. 1999;23(Suppl 6):S24–S29. doi: 10.1038/sj.ijo.0800939. [DOI] [PubMed] [Google Scholar]
- 45.Jastroch M., Hirschberg V., Klingenspor M. No mitochondrial uncoupling artefact is caused by expression of uncoupling protein 1 in a mammalian cell culture: a new system to study mitochondrial carrier proteins. The FASEB Journal. 2011;25(1_supplement) 1044.3-1044.3. [Google Scholar]
- 46.Jastroch M., Hirschberg V., Klingenspor M. Functional characterization of UCP1 in mammalian HEK293 cells excludes mitochondrial uncoupling artefacts and reveals no contribution to basal proton leak. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2012;1817(9):1660–1670. doi: 10.1016/j.bbabio.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Oelkrug R., Goetze N., Exner C., Lee Y., Ganjam G.K., Kutschke M. Brown fat in a protoendothermic mammal fuels eutherian evolution. Nature Communications. 2013;4 doi: 10.1038/ncomms3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu S., Jay A., Brunaldi K., Huang N., Hamilton J.A. CD36 enhances fatty acid uptake by increasing the rate of intracellular esterification but not transport across the plasma membrane. Biochemistry. 2013;52(41):7254–7261. doi: 10.1021/bi400914c. [DOI] [PubMed] [Google Scholar]
- 49.Rial E., González-Barroso M., Fleury C., Iturrizaga S., Sanchis D., Jiménez-Jiménez J. Retinoids activate proton transport by the uncoupling proteins UCP1 and UCP2. The EMBO Journal. 1999;18(21):5827–5833. doi: 10.1093/emboj/18.21.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomás P., Jiménez-Jiménez J., Zaragoza P., Vuligonda V., Chandraratna R.A., Rial E. Activation by retinoids of the uncoupling protein UCP1. Biochimica et Biophysica Acta. 2004;1658(1–2):157–164. doi: 10.1016/j.bbabio.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 51.Houten S.M., Wanders R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. Journal of Inherited Metabolic Disease. 2010;33(5):469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Himms-Hagen J. Brown adipose tissue thermogenesis: interdisciplinary studies. Federation of American Societies for Experimental Biology Journal: Official Publication of the Federation of American Societies for Experimental Biology. 1990;4(11):2890–2898. [PubMed] [Google Scholar]
- 53.Chouchani E.T., Kazak L., Jedrychowski M.P., Lu G.Z., Erickson B.K., Szpyt J. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature. 2016;532(7597):112–116. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills E.L., Pierce K.A., Jedrychowski M.P., Garrity R., Winther S., Vidoni S. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature. 2018;1 doi: 10.1038/s41586-018-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer A.W., Hoefig C.S., Abreu-Vieira G., de Jong J.M.A., Petrovic N., Mittag J. Leptin raises defended body temperature without activating thermogenesis. Cell Reports. 2016;14(7):1621–1631. doi: 10.1016/j.celrep.2016.01.041. [DOI] [PubMed] [Google Scholar]
- 56.Lundby A., Lage K., Weinert B.T., Bekker-Jensen D.B., Secher A., Skovgaard T. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Reports. 2012;2(2):419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aguer C., McCoin C.S., Knotts T.A., Thrush A.B., Ono-Moore K., McPherson R. Acylcarnitines: potential implications for skeletal muscle insulin resistance. The FASEB Journal. 2014;29(1):336–345. doi: 10.1096/fj.14-255901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCoin C.S., Knotts T.A., Adams S.H. Acylcarnitines—old actors auditioning for new roles in metabolic physiology. Nature Reviews Endocrinology. 2015;11(10):617–625. doi: 10.1038/nrendo.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dittenhafer-Reed K.E., Richards A.L., Fan J., Smallegan M.J., Siahpirani A.F., Kemmerer Z.A. SIRT3 mediates multi-tissue coupling for metabolic fuel switching. Cell Metabolism. 2015;21(4):637–646. doi: 10.1016/j.cmet.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahn B.-H., Kim H.-S., Song S., Lee I.H., Liu J., Vassilopoulos A. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimen H., Han M.-J., Yang Y., Tong Q., Koc H., Koc E.C. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49(2):304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finley L.W.S., Haas W., Desquiret-Dumas V., Wallace D.C., Procaccio V., Gygi S.P. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen B.K., Gupta R., Baldus L., Lyon D., Narita T., Lammers M. Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nature Communications. 2019;10(1):1055. doi: 10.1038/s41467-019-09024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou T., Chung Y.-H., Chen J., Chen Y. Site-specific identification of lysine acetylation stoichiometries in mammalian cells. Journal of Proteome Research. 2016;15(3):1103–1113. doi: 10.1021/acs.jproteome.5b01097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [36] partner repository with the dataset identifier PXD013056. The cold stress dataset is annotated under the ‘MD10’ keys and the room temperature experiment is annotated under the ‘OMDYS21-32’ keys.