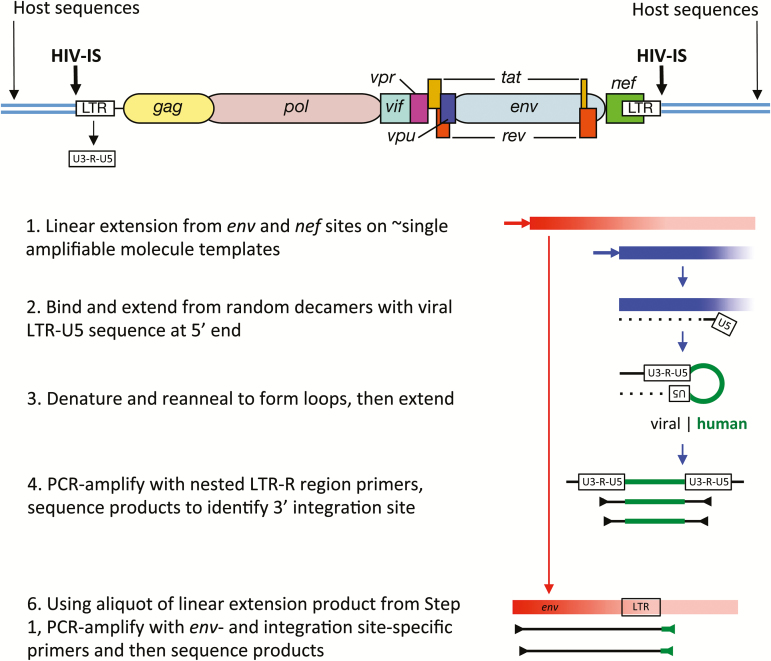
Figure 1.
Schematic summary of the Integration Site Looping Assay (ISLA). Essential aspects of this procedure are schematically shown here, whereas full details are provided in Supplementary Figure 1 from [31]. The structure of a full-length provirus and adjacent cellular sequences are shown at the top of the figure. Long terminal repeated sequences (LTRs) flank the provirus, with LTRs subdivided into U3, R, and U5 segments. These segment names derive from their origins in viral genomic RNA; they are found to be: unique to the 3’ end of the human immunodeficiency virus (HIV) genomic RNA (U3), repeated at each end of the HIV genomic RNA (R), and unique to the 5’ end of the HIV genomic RNA (U5). Using 2 virus-specific primers from the env and nef genes, linear amplification is carried out to increase the representation of these sequences in the cellular DNA preparation (Step 1). Products extending from the nef primer that reach the adjacent host sequences are much shorter (approximately 0.8kb vs approximately 2.8 kb) and hence are much more efficiently generated. An aliquot of this reaction is then used to extend DNA from random primer sequences containing the U5 region at the 5’ end (Step 2). This product is then denatured and reannealed to form a loop with the U5 region of the provirus hybridizing to its complement derived from extension products generated with random primers (Step 3). Such products are then selectively polymerase chain reaction (PCR)–amplified using R region primers and then sequenced to determine the host genome sequences immediately adjacent to the 3’ end of the provirus (ie, the integration site, Step 4). To link the integration site to a specific proviral genome, the sequence obtained needs to extend into the gp120 protein coding sequence of the viral env gene, the region of highest level of viral genetic variation. Hence, in step 5, integration site (IS)–specific primers are generated to provide the specificity to amplify the longer, less efficiently produced env IS products.