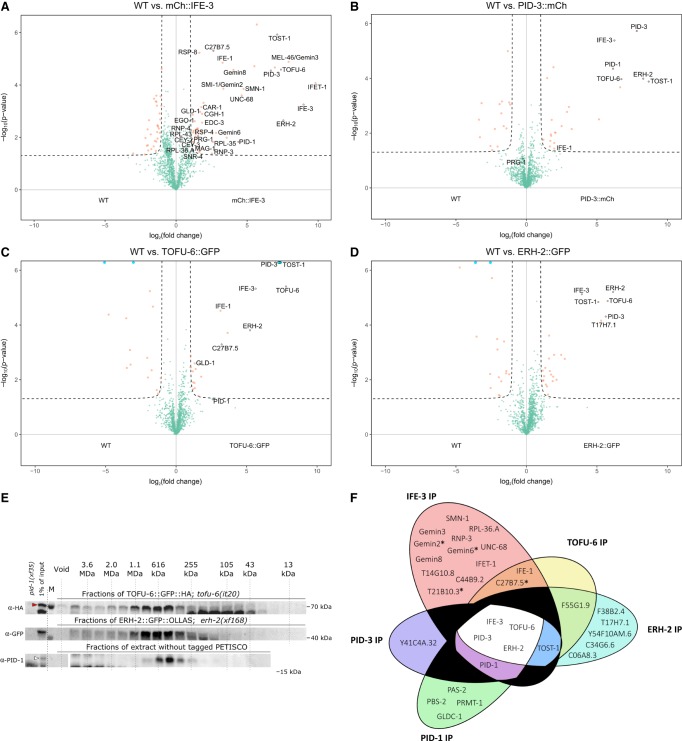
Figure 2.
PID-1 interactors form a novel protein complex, PETISCO. (A–D) Volcano plot representing label-free proteomic quantification of quadruplicate 3xFlag::mCherry::IFE-3;ife-3(xf101) (A), PID-3::mCherry::Myc;pid-3(tm2417) (B), TOFU-6::GFP::HA;tofu-6(it20) (C), and ERH-2::GFP::OLLAS;erh-2(xf168) (D) immunoprecipitations from nongravid adult extracts. The X-axis represents the median fold enrichment of individual proteins in the control (wild-type [WT]) versus the transgenic strain. The Y-axis indicates −log10(P-value) of observed enrichments. Dashed lines represent thresholds at P = 0.05 and twofold enrichment. Blue data points represent values out of scale. Red and green data points represent above and below threshold, respectively. (E) Size exclusion chromatography of nongravid adult worm extracts of the indicated strains. Fractions were collected and probed for HA, GFP, and PID-1, respectively. The approximate molecular weights of the fractions are indicated. pid-1(xf35) extract was used as probing control. The red arrowhead indicates full-length TOFU-6::GFP, and the white arrowhead indicates PID-1. pid-1(xf35) extract was used as negative control for PID-1. (F) Venn diagram summarizing significant interactions in PETISCO protein immunoprecipitations. The asterisk represents protein found significantly enriched in only one experiment of 3xFlag::mCherry::IFE-3;ife-3(xf101) immunoprecipitation.