Figure 1.
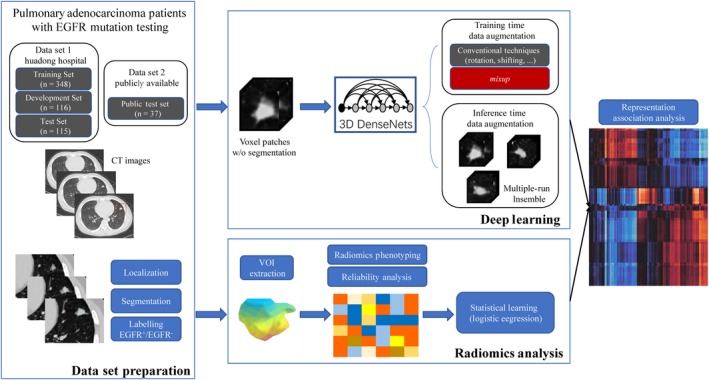
Overall pipeline for this study. A local CT dataset (HdH Dataset) and a public dataset selected from TCIA database (TCIA Dataset) of lung adenocarcinoma patients with EGFR mutation testing were used. Nodules were manually localized, segmented, and labelled as EGFR mutant (Mut)/wild‐type (WT). For deep learning, 3D DenseNets were trained using the training subset. A strong data augmentation technique, mixup, was used for better regularization. Expressive representations, that is, deep learned radiomics (DLR), for the nodules were end‐to‐end learned during the training procedure. Meanwhile, conventional radiomics analysis following the common practice was carried out for performance comparison, and association study between the 3D deep leaning and conventional radiomics was performed by calculating the pairwise correlation coefficients
