Abstract
In order to function on the ribosome with uniform rate and adequate accuracy, each bacterial tRNA has evolved to have a characteristic sequence and set of modifications that compensate for the differing physical properties of its esterified amino acid and its codon-anticodon interaction. The sequence of the T-stem of each tRNA compensates for the differential effect of the esterified amino acid on the binding and release of EF-Tu during decoding. The sequence and modifications in the anticodon loop and core of tRNA impact the codon-anticodon strength and the ability of the tRNA to bend during codon recognition. These discoveries impact the design of tRNAs for the efficient and accurate incorporation of unnatural amino acids into proteins using bacterial translation systems.
Introduction
When using the bacterial translational machinery to introduce unnatural amino acids (Uaas) into proteins, it is frequently assumed that tRNAs are generic adaptors that simply serve to physically connect the esterified amino acid and the anticodon and present them to the ribosome at the appropriate distance and orientation needed for catalysis. This “generic adaptor” model of tRNA function implies that any tRNA species could be chosen to acylate with a desired Uaa, and its anticodon could be altered to read any unassigned codon, such as a stop codon, a missense codon or even a codon containing non-natural nucleotides. This review will summarize experiments indicating that this generic adaptor model is not correct when tRNA function in translation is examined in greater detail and more quantitatively. Instead, the structure each tRNA species has evolved idiosyncratically to optimize the rate and accuracy of incorporation of its natural amino acid in response to its natural codons. In other words, each aa-tRNA participates in translation somewhat differently, where the esterified amino acid, the anticodon and the tRNA body each make unique kinetic and/or thermodynamic contributions to translational function. As a result, acylation of a tRNA with a non-cognate or unnatural amino acid or any alteration of its anticodon “mistunes” the tRNA and can result in reduced rate and/or accuracy of translation. This “unique participant” model of tRNA function places important restrictions on the choice of tRNAs suitable for the efficient introduction of Uaas into proteins, especially when multiple insertions are desired. Additional reviews of this topic have recently appeared [1,2].
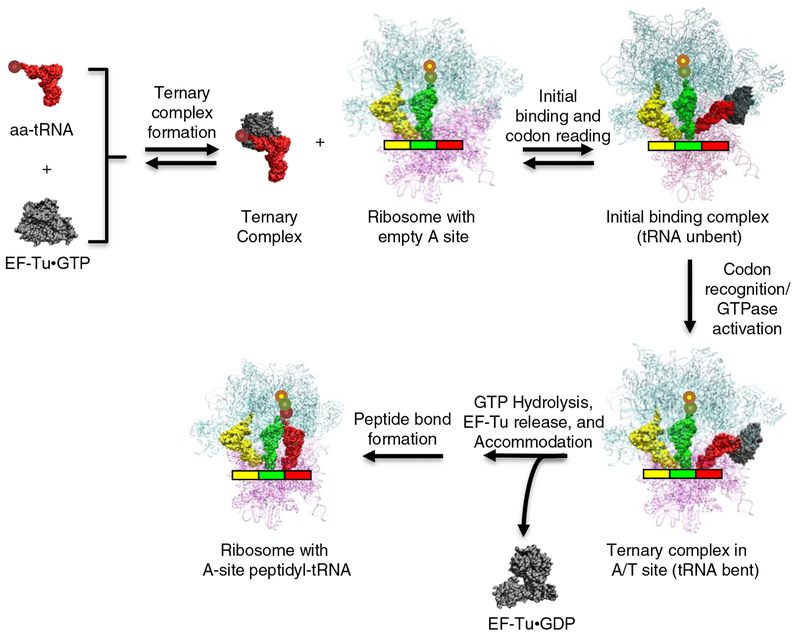
The steps in the translation cycle leading up to peptide bond formation are currently quite well understood both kinetically and structurally (Figure 1).From the perspective of the aa-tRNA substrate, decoding can be considered as two processes that occur at spatially distinct parts of the molecule. One process is the binding and subsequent release of aa-tRNA from EF-Tu, which involves contacts with the esterified amino acid and the acceptor and T helices. The other process is the bending of tRNA that accompanies codon binding and the subsequent unbending upon A-site entry, which involves the anticodon hairpin and much of the tertiary core of tRNA. An important conclusion of this review is that in order for the many different elongator tRNA species to undergo decoding at roughly similar rates and with optimal accuracy, each one has evolved to have different sequences and a unique pattern of nucleotide modifications. In the first process, tRNAs have evolved to compensate for the intrinsically different physical properties of their esterified amino acids. In the second process, tRNAs have evolved to compensate for the intrinsically differing strengths of their codon-anticodon interactions.
Figure 1. Steps in the mechanism of bacterial translational decoding.
For a full description of the decoding mechanism and the values for the rate constants of individual steps, see Rodnina et al. [48]. High resolution x-ray crystal structures are available for each of the ribosome bound intermediates [39,50,51] except for the labile initial binding complex. Structural diagrams were generated using VMD [52].
Evolutionary tuning to optimize the affinity between aa-tRNAs and EF-Tu
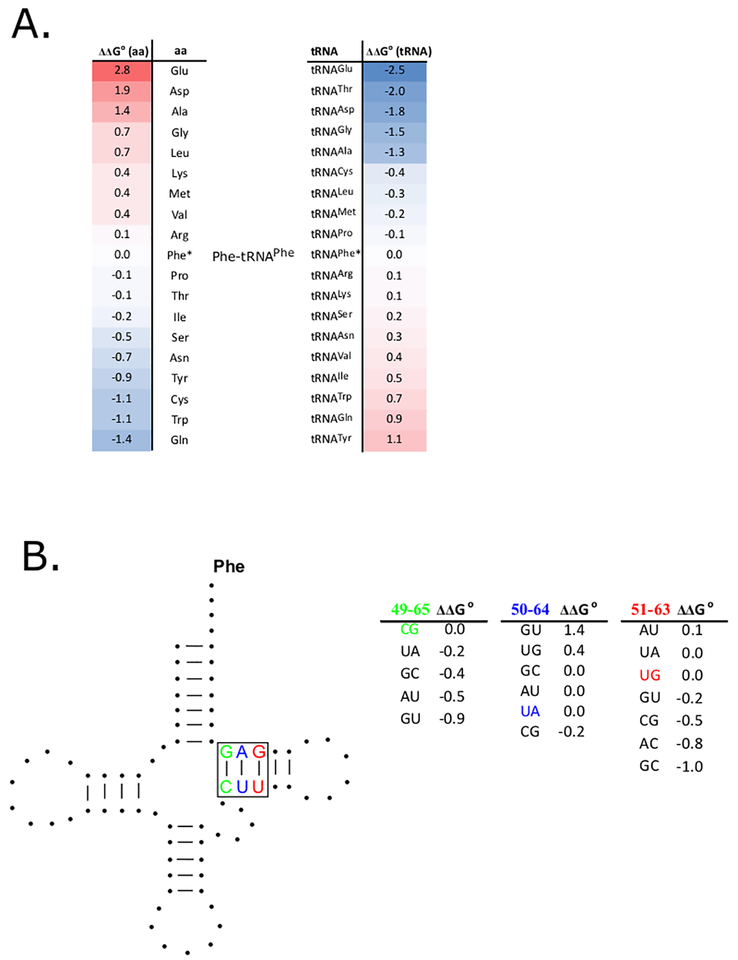
Consistent with their interchangeable function in translation, all the different native aa-tRNAs bind EF-Tu with similar affinities [3]. However, such uniform binding is not observed when the tRNAs are misacylated. Numerous experiments with misacylated tRNAs [4–6] support a simple model where the total free energy of binding of an aa-tRNA to EF-Tu (ΔG° (total)) can be described by independent free energy contributions of the esterified amino acid (ΔG°(aa)) and the tRNA body (ΔG°(tRNA)) (Figure 2A). The energetic contributions of different amino acids and different tRNA bodies both vary significantly and the values generally offset one another for tRNAs esterified with their cognate amino acids.
Figure 2. Structural elements in aa-tRNAs that contribute to EF-Tu binding specificity.
A. Variable contributions of amino acid and tRNA body to EF-Tu affinity. ΔΔG° values for substituting either the amino acid (left) or the tRNA body (right) of Phe-tRNAPhe are listed. Both substitutions are ranked from the most stabilizing (blue) to the most destabilizing (red), but are presented in inverse order to emphasize that the values compensate for correctly acylated tRNAs [6]. By using a ΔG°(total) = −10.1 kcal/mol for Phe-tRNAPhe, the ΔG°(total) of any misacylated aa-tRNA can be calculated from these data [5]. Values are only appropriate for the experimental conditions used in [5] since EF-Tu binding affinity is very dependent on temperature and ionic strength.
B. T-stem sequence affects aa-tRNA binding to EF-Tu. ΔΔG° values for substituting the C49-G65 (green), U50-A64(blue) and U51-G63 (red) pairs present in E. coli tRNAPhe for other base pairs. Since these ΔΔG° values contribute independently from one another, these data can be used to calculate the ΔG°(tRNA) for any tRNA sequence [13] and combined with data in panel A to estimate ΔG°(total) for any aa-tRNA.
Crystal structures of ternary complexes with either an esterified Phe or Cys [7,8] show that the side chain of the amino acid fits into a pocket in EF-Tu and is stabilized by stacking upon His66 that lies in the base of the pocket. Although this pocket is large enough to fit all the natural amino acids, it is currently not known exactly how the other amino acids are accommodated. However, the irregular shape and overall negative charge of the pocket is consistent with each amino acid having a different ΔG°(aa). As expected, EF-Tu mutations of His66 [9] and other residues that line the pocket result in substantial changes in the specificity for different amino acids compared to the wild type protein.
The extensive interface between EF-Tu and tRNA involves multiple thermodynamically significant contacts between the coaxially stacked acceptor and T stems of tRNA and 22 amino acids in domains 1 and 3 of EF-Tu [10,11]. However, tRNA mutagenesis experiments show that most of the variation in ΔG°(tRNA) is the result of only part of the interface involving three adjacent base pairs in the T-stem [12]. The contribution of each of the base pairs to ΔG°(tRNA) is independent of the other two and each base pair shows its own characteristic sequence dependence. By combining EF-Tu binding data using sets of T stem mutations made in three different tRNAs, an empirical thermodynamic “code” for predicting ΔG° (tRNA) for any T stem sequence was deduced (Figure 2B). When this code was applied to almost 6000 different bacterial tRNAs, the ΔG°(tRNA) for each tRNA species was predicted to fall within a narrow range, despite the fact that their T stem sequences sometimes varied dramatically [13]. For example, although the tRNAThrUGU from a set of 158 bacteria had 53 different T stem sequences, they all had a similar calculated value of ΔG°(tRNA). The different T stem sequences observed for a given tRNA in different bacteria therefore represent alternative evolutionary “solutions” to obtain its characteristic ΔG° (tRNA).
Crystal structures of isolated ternary complexes [7,8] as well as ribosome bound ternary complexes either just before [14] or just after [15] GTP hydrolysis show that the interface between aa-tRNA and EF-Tu remains virtually unchanged throughout decoding. This suggests that the rate of release of aa-tRNAs from EF-Tu on the ribosome may also be subject to a similar dependence on the identity of the esterified amino acid and tRNA body. This expectation was confirmed by assaying a set of T-stem mutations and misacylated versions of tRNAvalGAC that either tightened or weakened ΔG° (total) [16]. As expected, the weaker binding variants functioned poorly due to incomplete ternary complex formation. In contrast, the tighter binding variants bound ribosomes well, but showed decoding rates that were reduced in precise proportion to their binding affinities, reflecting their slower release from EF-Tu after GTP hydrolysis. Thus, the value of ΔG° (total) has been “tuned” by evolution to have an optimal value. If ΔG° (total) is too weak, the aa-tRNA does not bind EF-Tu and cannot enter protein synthesis. If ΔG° (total) is too tight, the aa-tRNA does not release from EF-Tu fast enough during decoding and the rate of subsequent peptide bond formation decreases.
The discovery that the identity of the esterified amino acid contributes substantially to EF-Tu affinity has important implications in choosing an appropriate tRNA to incorporate a given Uaa. Ideally, an Uaa-tRNA should have a ΔG°(total) similar to the value typical for all correctly acylated native aa-tRNAs. Although Uaas with tight ΔG°(aa)s are possible, ΔG°(aa) of most Uaas studied thus far are quite weak compared to natural amino acids, resulting in a lower fraction of the Uaa-tRNA forming ternary complex and slower incorporation into peptide [17–19]. For several Uaas with weak ΔG°(aa)s, incorporation efficiency can be improved by either increasing EF-Tu concentration[18], choosing a tRNA with a tighter ΔG°(tRNA) [17,20], or modifying the sequence of the T-stem to strengthen ΔG°(tRNA) [20–22]. An alternative approach for improving weak ΔG°(aa) values is to mutate the residues that form the amino acid binding pocket of EF-Tu. Although ΔG°(aa) of several bulky Uaas were improved by mutations designed to enlarge the pocket [23], incorporation yields remained low compared to native aa-tRNAs. A careful kinetic analysis of one bulky Uaa-tRNA showed that while the amino acid pocket mutations strengthened binding to EF-Tu, peptide bond formation remained quite slow suggesting that the mutations had also compromised some step after GTP hydrolysis [19]. This may reflect an additional step in decoding that has recently been proposed [24]. However, the successful selection of EF-Tu pocket mutations that improved both Uaa-tRNA binding and subsequent incorporation of phosphoserine [25], phosphotyrosine [26] and p-azidophenylalanine [27] suggests that the development of EF-Tu molecules that are specific for a given Uaa will be beneficial.
Evolutionary tuning of tRNAs to optimize codon recognition
In the same way that the T stem sequence has been tuned in response to the side chain of the esterified amino acid, other structural elements in each tRNA species have been idiosyncratically tuned by evolution to ensure both efficient and accurate codon recognition. Since codon recognition requires both the formation of a sufficiently stable codon-anticodon complex and the ability to achieve a bent conformation, both of these features will influence its rate. The stabilities of the 61 different codon-anticodon complexes are expected to vary substantially due to their varied sequence and GC content. Similarly, one would expect that the ability of different tRNAs to bend appropriately will depend on the sequence of the region that bends. The thermodynamic interplay between codon binding strength and tRNA bending is critical for the decoding process. If the codon-anticodon interaction is too weak or the tRNA is unable to bend easily enough (too “stiff”), then the ternary complex will not remain bound in the A/T-site long enough for GTP hydrolysis to proceed. If the codon-anticodon interaction is too tight or the tRNA bends too easily (too “flexible”), near-cognate codons could function well enough for GTP hydrolysis and peptide bond formation to proceed, leading to misreading. Thus, for each tRNA species, the anticodon has coevolved with structural elements in the anticodon hairpin and tRNA body to achieve the stability and flexibility necessary for uniform and accurate codon recognition.
Although less complete than the data for the much simpler EF-Tu tuning, there is abundant experimental evidence that the structures of different tRNAs are also idiosyncratically tuned by evolution to optimize the codon recognition process. Initial experiments focused on the unique pattern of modifications present on each tRNA. Indeed, it was observed almost 50 years ago that the type of modification present at position 37 correlated with the identity of the adjacent anticodon residue 36 [28]. When the modification of position 37 was removed or altered, decoding efficiency and/or accuracy was compromised [29,30]. Although beyond the scope of this review, other idiosyncratic modifications at anticodon positions 34 and 35 can also stabilize codon binding and even modulate base pairing specificity [31]. In addition to the modifications, there are many anecdotal examples where structurally conservative sequence changes in the anticodon hairpin or the tertiary core of tRNA affect codon recognition efficiency and/or accuracy. Changing base pairs at several positions in the anticodon stem and the tertiary core of tRNA significantly modulates decoding efficiency [32–35]. The rare A32-U38 pair present in tRNAAlaGGC acts to weaken ribosome binding and thereby prevents very efficient misreading [36,37]. The G24A mutation in the D helix of tRNATrpCCA strongly promotes misreading [38], and a crystal structure revealed that the mutant stabilized the bent state by forming an additional hydrogen bond [39]. Finally, an extensive mutagenic study of tRNAAlaGGC identified many structurally conservative mutations that promoted significant misreading and occurred precisely in the region of the catalytic core known to bend [40]. Thus, it is clear that both the sequence and the modifications of each tRNA species act together to optimize codon recognition. However no “code” is yet available to predict how this works across tRNA species. The comparison of the structures of each tRNA species in many different bacteria should provide a promising starting point for deriving such a code. However, although a clear “consensus” sequence for each tRNA species has been defined [41], our knowledge of their corresponding tRNA modifications in multiple bacteria is not yet available [42].
The fact that the sequence and modifications on each tRNA body have coevolved with the anticodon to optimize the efficiency and accuracy of codon recognition complicates the use of anticodon substituted tRNAs for the introduction of Uaas. If a tRNA body has an inappropriate anticodon, the resulting “mistuned” Uaa-tRNA may either progress less quickly through the decoding steps or function too well and insert Uaas at incorrect codons. Although many kinds of anticodon substituted tRNAs have been used for insertion of Uaas [43,44], a common strategy is to use different amber suppressor tRNAs (SutRNAs) where one or more anticodon residues of an elongator tRNA are mutated to read the unassigned UAG codon. SutRNAs are mistuned since the sequences of their anticodon hairpins and tRNA bodies evolved to decode their original codons and not the UAG codon. In addition, the modifications present on many SutRNAs are not only inappropriate for the UAG codon, but are often not present since many of the anticodon loop modifying enzymes require the correct anticodon sequence to function. Although no SutRNA has been subjected to careful kinetic analysis, most, if not all, undergo decoding less well than elongator tRNAs [Box 1]. Several of the more efficient SutRNAs can successfully introduce a single Uaa yet their compromised activity is evident by their low efficiency of incorporation of multiple Uaas. Although the development of E. coli strains missing the competing RF1 protein has dramatically improved the efficacy of SutRNAs [45], identifying a SutRNA that is fully tuned to read the UAG codon remains an important goal. While the rare tRNAPyl naturally uses a UAG codon, it functions poorly and thus must be mistuned in some way [46]. Mutagenizing tRNAPyl [47] or SutRNAs [22] may improve tuning. Once candidate tuned SutRNAs are identified, it will be important to carefully measure the multiple rate constants that describe decoding [48] using both the cognate UAG and several near-cognate codons in order to ensure that they have rates and accuracies similar to normal elongator tRNAs.
Box 1: Evaluating mistuned Uaa-tRNAs.
The efficacies of Uaa-tRNAs are often evaluated by comparing the yield of a test protein containing a single Uaa with the yield of a native protein control reaction. However, this criterion does not accurately evaluate Uaa-tRNA function because it measures the overall yield of the multiple turnover, multistep in vitro translation reaction instead of measuring the rate of the Uaa insertion step. The rate of synthesis of a complete protein is only modestly affected by the rate of an individual step. For example, using a typical average step time of 250ms, the elongation of a 300 aa protein requires about 15s. However, a badly “mistuned” Uaa-tRNA that decodes 100 times slower would take 5s to be inserted. Thus, a single insertion would only increase the time to complete the protein from 15 to 20s, which will be hard to detect in the overall yield of protein in the lengthy, multiple turnover reactions that are typically used. To properly evaluate an Uaa-tRNA, the time required for a single incorporation event must be measured directly. A fully tuned Uaa-tRNA should approach the 50ms incorporation time typical for native aa-tRNAs
Even when proteins with only a single Uaa are desired, lengthy ribosomal “stalling” at the incorporation site of a mistuned Uaa-tRNA can lead to several undesirable side reactions:
Increased error rates due to misincorporation by a near-cognate aa-tRNA if the A-site is empty or frameshifting if P-site entry is slow.
Premature termination due to competition by termination factors if nonsense suppressor tRNAs are used.
Significant rates of spontaneous peptidyl-tRNA dissociation when nascent chains are short [53]
Spontaneous deacylation of the valuable (and often limiting) Uaa-tRNA that is especially rapid when not bound to EF-Tu.
In summary, although our understanding of how the structures of individual bacterial tRNAs influence their ribosomal decoding properties remains rudimentary (Figure 3), it is clear that each tRNA decodes differently. Although experimental data is sparse [49], a similar conclusion may be true for one or more of the subsequent steps describing the passage of tRNA through the ribosomal P and E-sites. Thus, the design of optimized Uaa-tRNAs will continue to be challenging.
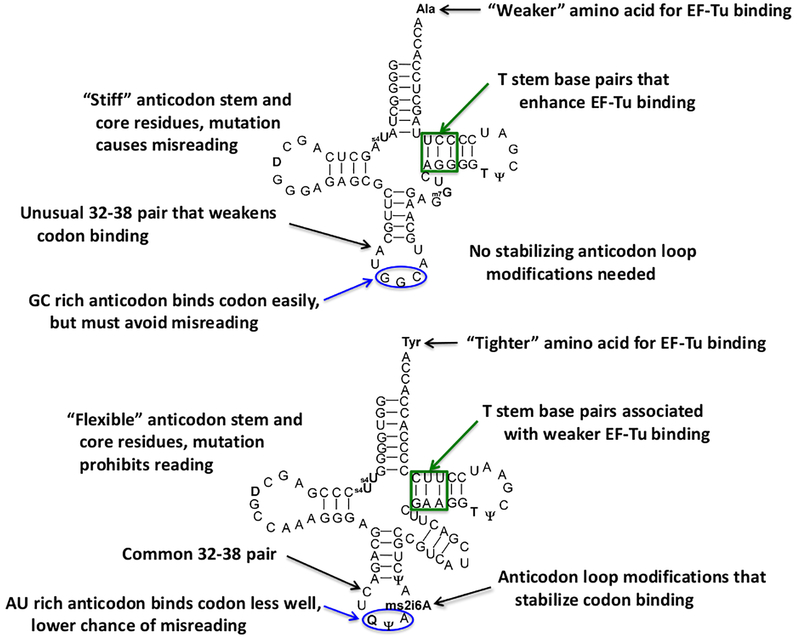
Figure 3. Idiosyncratic evolutionary tuning of two E coli tRNAs.
Two tRNAs with quite different decoding strategies are compared. tRNAAlaGGC possesses an amino acid that binds EF-Tu weakly and has a very stable codon-anticodon interaction. tRNATyrGUA possesses a tighter binding amino acid and a weaker codon-anticodon interaction. While EF-Tu binding data for both tRNAs is well established, systematic mutagenesis of the anticodon stem and core has only been performed for tRNAAlaGGC [40]. Since no direct measurement of the flexibility of these tRNAs has been performed, these descriptions are speculative. Modified nucleotides are in bold [42], the EF-Tu recognition region is boxed in green and the anticodon is circled in blue.
Acknowledgements
This work was supported by NIGMS of the National Institutes of Health under award number R35GM124733 to JMS and Wayne State University startup funds to JMS.
References
- 1.Reynolds NM, Vargas-Rodriguez O, Soll D, Crnkovic A: The central role of tRNA in genetic code expansion. Biochim Biophys Acta 2017, 1861:3001–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katoh T, Iwane Y, Suga H: tRNA engineering for manipulating genetic code. RNA Biol 2017:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louie A, Ribeiro NS, Reid BR, Jurnak F: Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem 1984, 259:5010–5016. [PubMed] [Google Scholar]
- 4.Asahara H, Uhlenbeck OC: The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci U S A 2002, 99:3499–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asahara H, Uhlenbeck OC: Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu.GTP. Biochemistry 2005, 44:11254–11261. [DOI] [PubMed] [Google Scholar]; * 5. A comprehensive view of how the esterified amino acid and tRNA body independently contribute to EF-Tu binding affinity.
- 6.LaRiviere FJ, Wolfson AD, Uhlenbeck OC: Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 2001, 294:165–168. [DOI] [PubMed] [Google Scholar]
- 7.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J: Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 1995, 270:1464–1472. [DOI] [PubMed] [Google Scholar]
- 8.Nissen P, Thirup S, Kjeldgaard M, Nyborg J: The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 1999, 7:143–156. [DOI] [PubMed] [Google Scholar]
- 9.Chapman SJ, Schrader JM, Uhlenbeck OC: Histidine 66 in Escherichia coli elongation factor tu selectively stabilizes aminoacyl-tRNAs. J Biol Chem 2012, 287:1229–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanderson LE, Uhlenbeck OC: Directed Mutagenesis Identifies Amino Acid Residues Involved in Elongation Factor Tu Binding to yeast Phe-tRNA(Phe). J Mol Biol 2007, 368:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yikilmaz E, Chapman SJ, Schrader JM, Uhlenbeck OC: The interface between Escherichia coli elongation factor Tu and aminoacyl-tRNA. Biochemistry 2014, 53:5710–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrader JM, Chapman SJ, Uhlenbeck OC: Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol 2009, 386:1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrader JM, Uhlenbeck OC: Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria? Nucleic Acids Res 2011, 39:9746–9758. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** 13. An empirical model permits the accurate calculation of ΔG°(tRNA) for nearly 6000 bacterial tRNAs based on their T stem sequences. The model explains the variability in T stem sequences for a given tRNA species among different bacteria and correctly predicts weak EF-Tu binding for aa-tRNAs known to not participate in elongation.
- 14.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V: The mechanism for activation of GTP hydrolysis on the ribosome. Science 2010, 330:835–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V: The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 2009, 326:688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrader JM, Chapman SJ, Uhlenbeck OC: Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci U S A 2011, 108:5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ieong KW, Pavlov MY, Kwiatkowski M, Ehrenberg M, Forster AC: A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids. RNA 2014, 20:632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** 17. Elegant rapid reaction methods show that biphasic progress curves of peptide bond formation for several different Uaa-tRNAPhes are due to inefficient EF-Tu binding and fast rates of peptide bond formation. The tighter binding tRNAAla body increases the overall rate of Uaa incorporation by increasing the proportion of Uaa-tRNAs bound to EF-Tu.
- 18.Ieong KW, Pavlov MY, Kwiatkowski M, Forster AC, Ehrenberg M: Inefficient delivery but fast peptide bond formation of unnatural L-aminoacyl-tRNAs in translation. J Am Chem Soc 2012, 134:17955–17962. [DOI] [PubMed] [Google Scholar]
- 19.Mittelstaet J, Konevega AL, Rodnina MV: A kinetic safety gate controlling the delivery of unnatural amino acids to the ribosome. J Am Chem Soc 2013, 135:17031–17038. [DOI] [PubMed] [Google Scholar]
- 20.Achenbach J, Jahnz M, Bethge L, Paal K, Jung M, Schuster M, Albrecht R, Jarosch F, Nierhaus KH, Klussmann S: Outwitting EF-Tu and the ribosome: translation with d-amino acids. Nucleic Acids Res 2015, 43:5687–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]; * 20 The incorporation of many D-amino acids was improved using the tight EF-Tu binding tRNAGly.
- 21.Katoh T, Iwane Y, Suga H: Logical engineering of D-arm and T-stem of tRNA that enhances d-amino acid incorporation. Nucleic Acids Res 2017, 45:12601–12610. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** 21 The normally poor incorporation of D amino acids is dramatically improved by engineering the T stem of tRNAPro to bind EF-Tu more tightly and by including elongation factor P in the translation reaction.
- 22.Guo J, Melancon CE 3rd, Lee HS, Groff D, Schultz PG: Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids. Angew Chem Int Ed Engl 2009, 48:9148–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M: Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J Am Chem Soc 2007, 129:14458–14462. [DOI] [PubMed] [Google Scholar]
- 24.Ieong KW, Uzun U, Selmer M, Ehrenberg M: Two proofreading steps amplify the accuracy of genetic code translation. Proc Natl Acad Sci U S A 2016, 113:13744–13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, Soll D: Expanding the genetic code of Escherichia coli with phosphoserine. Science 2011, 333:1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan C, Ip K, Soll D: Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett 2016, 590:3040–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan R, Perez JG, Carlson ED, Ntai I, Isaacs FJ, Kelleher NL, Jewett MC: Translation system engineering in Escherichia coli enhances non-canonical amino acid incorporation into proteins. Biotechnol Bioeng 2017, 114:1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura S: Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol 1972, 12:49–85. [PubMed] [Google Scholar]
- 29.Manickam N, Joshi K, Bhatt MJ, Farabaugh PJ: Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res 2016, 44:1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agris PF, Eruysal ER, Narendran A, Vare VYP, Vangaveti S, Ranganathan SV: Celebrating wobble decoding: Half a century and still much is new. RNA Biol 2017:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vare VY, Eruysal ER, Narendran A, Sarachan KL, Agris PF: Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raftery LA, Yarus M: Systematic alterations in the anticodon arm make tRNA(Glu)-Suoc a more efficient suppressor. EMBO J 1987, 6:1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarus M: Translational efficiency of transfer RNA’s: uses of an extended anticodon. Science 1982, 218:646–652. [DOI] [PubMed] [Google Scholar]
- 34.Schultz DW, Yarus M: tRNA structure and ribosomal function. I. tRNA nucleotide 27–43 mutations enhance first position wobble. J Mol Biol 1994, 235:1381–1394. [DOI] [PubMed] [Google Scholar]
- 35.Schultz DW, Yarus M: tRNA structure and ribosomal function. II. Interaction between anticodon helix and other tRNA mutations. J Mol Biol 1994, 235:1395–1405. [DOI] [PubMed] [Google Scholar]
- 36.Ledoux S, Olejniczak M, Uhlenbeck OC: A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol 2009, 16:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami H, Ohta A, Suga H: Bases in the anticodon loop of tRNA(Ala)(GGC) prevent misreading. Nat Struct Mol Biol 2009, 16:353–358. [DOI] [PubMed] [Google Scholar]
- 38.Cochella L, Green R: An active role for tRNA in decoding beyond codon:anticodon pairing. Science 2005, 308:1178–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V: How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol 2011, 18:432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]; * 39 Crystal structures of tRNAs trapped in their bent state during decoding are used to explain how certain mutants in the body of tRNATrp promote misreading.
- 40.Shepotinovskaya I, Uhlenbeck OC: tRNA residues evolved to promote translational accuracy. RNA 2013, 19:510–516. [DOI] [PMC free article] [PubMed] [Google Scholar]; **40 A comprehensive analysis of structurally conservative mutations in the anticodon stem and body of E coli tRNAAla shows that efficient misreading occurs with mutants in the region of tRNA expected to bend. Only mutations that did not fit the “consensus” tRNAAla sequence derived from comparing bacterial genomes showed misreading.
- 41.Saks ME, Conery JS: Anticodon-dependent conservation of bacterial tRNA gene sequences. RNA 2007, 13:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. : MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW: Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 2010, 464:441–444. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Ptacin JL, Fischer EC, Aerni HR, Caffaro CE, San Jose K, Feldman AW, Turner CR, Romesberg FE: A semi-synthetic organism that stores and retrieves increased genetic information. Nature 2017, 551:644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L: RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol 2011, 7:779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Kwiatkowski M, Forster AC: Kinetics of tRNA(Pyl) -mediated amber suppression in Escherichia coli translation reveals unexpected limiting steps and competing reactions. Biotechnol Bioeng 2016, 113:1552–1559. [DOI] [PubMed] [Google Scholar]
- 47.Fan C, Xiong H, Reynolds NM, Soll D: Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res 2015, 43:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodnina MV, Fischer N, Maracci C, Stark H: Ribosome dynamics during decoding. Philos Trans R Soc Lond B Biol Sci 2017, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fei J, Richard AC, Bronson JE, Gonzalez RL Jr.: Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol 2011, 18:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korostelev A, Trakhanov S, Laurberg M, Noller HF: Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 2006, 126:1065–1077. [DOI] [PubMed] [Google Scholar]
- 51.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V: Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K: VMD: visual molecular dynamics. J Mol Graph 1996, 14:33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Kwiatkowski M, Forster AC: Kinetics of Ribosome-Catalyzed Polymerization Using Artificial Aminoacyl-tRNA Substrates Clarifies Inefficiencies and Improvements. ACS Chem Biol 2015, 10:2187–2192. [DOI] [PubMed] [Google Scholar]