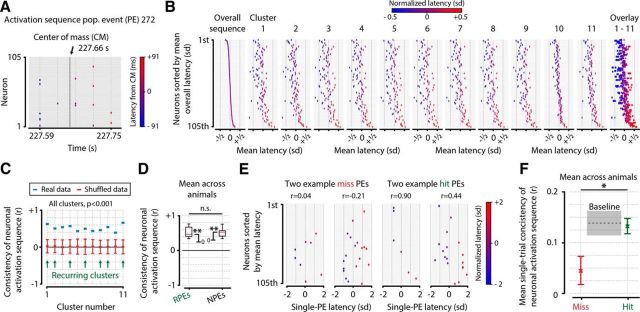
Figure 8.
Neurons show consistent, preferred temporal locations of activity within recurring and NPEs, and this temporal positioning is more precise during stimulus detection. A, For each PE (panel shows an example event), we calculated the mean activation latency (in milliseconds) per neuron to the center of mass of the event. B, Example sequences of activation for all clusters averaged across the cluster's PEs, ordered by position in the overall sequence. C, Quantification of B, showing that, across all clusters, including both recurring and nonrecurring events, the temporal sequence of activation is consistent, and significantly more so than expected by chance (permutation test vs 0, n = 11, for all clusters, p < 0.001). D, Mean across animals (n = 8) of sequence consistency was significantly >0, for both recurring clusters (mean consistency ± SEM, r = 0.505 ± 0.06, t test vs 0, n = 8, p = 5.37 × 10−5) and nonrecurring clusters (mean ± SEM, r = 0.497 ± 0.05, p = 1.71 × 10−5). The difference in consistency between recurring and nonrecurring events was not significant (paired t test, n = 8, p = 0.924, not significant). This suggests that the preferred temporal position of a neuron's activation is not related to the specific configuration of active neurons but is rather a universal property of all PEs. E, F, Single PE, rather than cluster-based sequence analyses. E, Four example PEs during stimulus presentation of test contrasts (0.5%–32%), showing correlation values with the overall sequence. The displayed events were chosen to illustrate the difference between high and low sequence consistency and are therefore not representative of the actual hit/miss effect size. F, Quantification of average single-trial sequence consistency during misses (red), hits (green), and preceding stimulus presentation (gray), showing that population sequences of neuronal activity are more temporally consistent when the visual stimulus is behaviorally reported (paired t test of miss vs hit consistency, n = 8 animals, p = 0.022). Colored error bars and gray baseline indicate mean ± SEM across animals. *, p < 0.05; **, p < 10−4 n.s., Not significant.