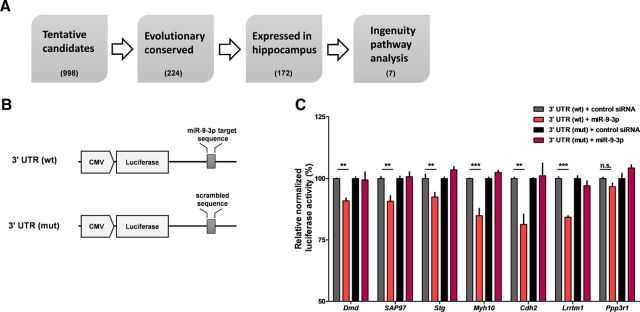
Figure 5.
miR-9-3p regulates the expression of LTP-related genes. A, Flow chart of bioinformatic analyses. The TargetScan algorithm predicted tentative target genes of miR-9-3p. Further analyses were performed based on evolutionary conservation of miR-9-3p target sequence and reliable expression in hippocampus. Finally, seven candidate target genes were identified via Ingenuity Pathway Analysis. Numbers in parentheses indicate the number of counted target genes via each bioinformatics analysis. B, Schematic diagrams of the reporter constructs used for the luciferase assays. C, miR-9-3p suppresses the expression of the luciferase reporters containing the 3′ UTRs of selected LTP-related genes [n = 4–6 for each group, 2-way ANOVA, miR-9-3p × mutation; Dmd, F(1,20) = 5.91, p = 0.0245; SAP97, F(1,20) = 9.59, p = 0.0057; Stg, F(1,20) = 14.97, p = 0.001; Myh10, F(1,20) = 31.39, p < 0.0001; Cdh2, F(1,12) = 8.95, p = 0.0112; Lrrtm1, F(1,12) = 27.5, p = 0.0002; Ppp3r1, F(1,12) = 7.06, p = 0.0209; Bonferroni's post tests, 3′ UTR (wt) + control siRNA versus 3′ UTR (wt) + miR-9-3p, **p < 0.01, ***p < 0.001]. Data are mean ± SEM.