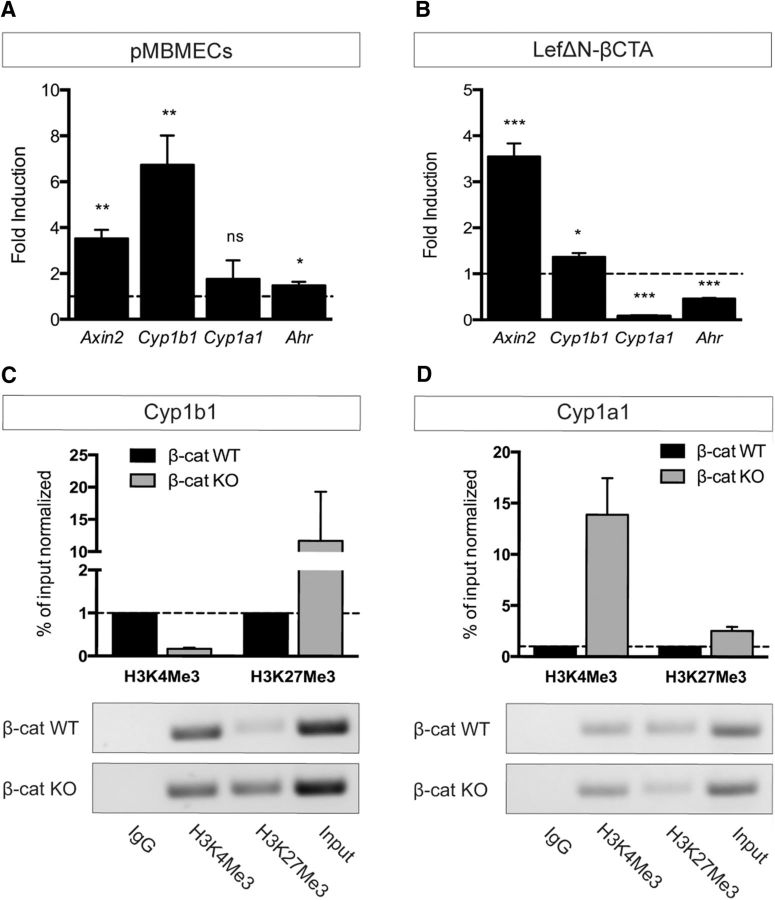
Figure 2.
β-Catenin transcriptional activity oppositely regulates Cyp1b1 and Cyp1b1 expression in ECs. A, qRT-PCR of pMBMECs stimulated with Wnt3a CM for 5 d relative to control CM (dashed line, n = 4). B, mRNA expression of Axin2, Cyp1b1, Cyp1a1, and Ahr in β-catenin KO ECs transduced with LefΔN-βCTA relative to the vector control (dashed line, n = 3). *p < 0.05. **p < 0.01. ***p < 0.001. ns, Not significant. Error bars indicate SEM. C, D, ChIP analysis of β-catenin WT and KO ECs determining the enrichment of either activating (trimethylation of histone H3 at lysine 4: H3K4Me3) or repressive (trimethylation of histone H3 at lysine 27: H3K27Me3) chromatin marks in Cyp1b1 (C) and Cyp1a1 (D) promoter regions. Quantitative ChIP-qPCR data are calculated as percentage over input normalized to β-catenin WT condition, which is assumed as 1 (dashed line). Columns represent mean ± SEM of two ChIP experiments. PCR products were visualized on agarose gels for qualitative analysis.