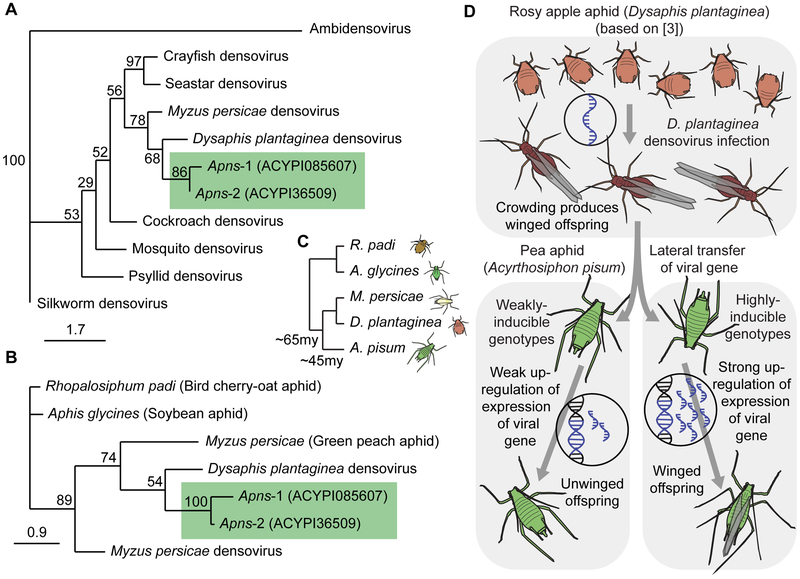
Figure 3: Origin of pea aphid densovirus genes and their retention of function.
A: Unrooted, bootstrap consensus tree showing the protein phylogenetic relationships among densovirus sequences from different invertebrate hosts, related viruses (parvovirus and ambidensovirus), and pea aphid Apns-1 and Apns-2. B: Unrooted, bootstrap consensus tree created from amino acid directed nucleotide alignments showing the phylogenetic relationships among densovirus sequences (from Myzus persicae and Dysaphis plantaginea) and aphid homologs (Rhopalosiphum padi, Aphis glycines, Myzus persicae, and pea aphid Apns-1 and Apns-2). For both trees, only nucleotide sequence alignable with the shorter pea aphid copy (Apns-1) were used. 1000 bootstrap trees were generated with maximum likelihood. C: Species tree for the aphid species referred to in Figure 3B. The relationships among species are inferred from trees in Kim et al. [32] and Hardy et al. [33]. Divergence times are based on [32]. D: Model illustrating densovirus gene domestication and subsequent retention of function. See Figure S3 and Table S3 for more information.