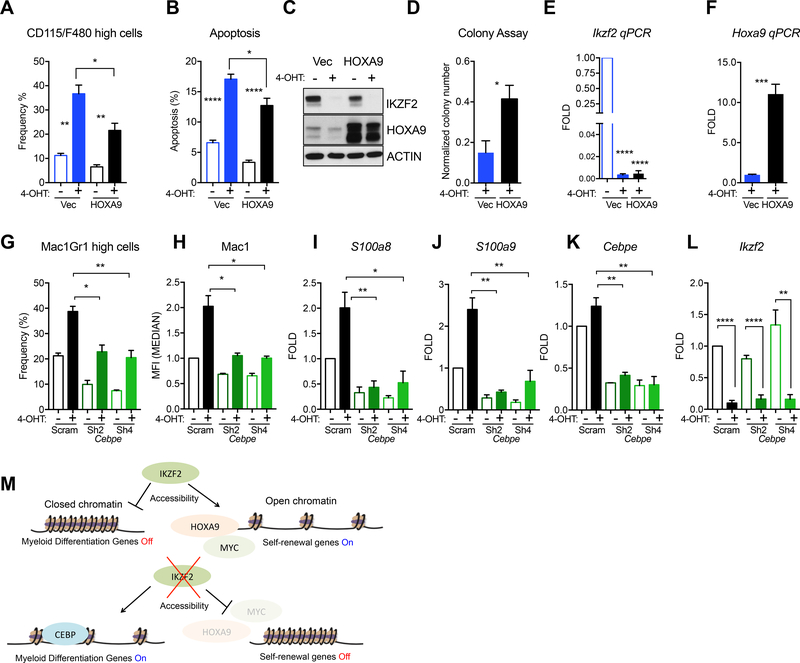
Figure 7. CEBPE and HOXA9 can partially rescue phenotypes of Ikzf2 loss.
(A) Differentiation measured by CD115/F480 and (B) apoptosis were partially rescued when Ikzf2 was deleted in HOXA9 overexpressing MLL-AF9 Ikzf2f/f cre-ER cells. n=3 independent experiments. Student’s t test *p<0.05, ** p<0.01, **** p<0.0001. (C) Western blot analysis demonstrating efficient deletion of IKZF2 and overexpression of HOXA9. (D) Colony assay showing rescue of Ikzf2 deletion by HOXA9 overexpression in 4-OHT treated MLL-AF9 Ikzf2f/f cre-ER cells. (E) QPCR showing complete deletion of Ikzf2 in MLL-AF9 Ikzf2f/f cre-ER vector control and HOXA9 overexpressing cells after 4-OHT treatment. (F) QPCR showing expression of Hoxa9 in the vector control and HOXA9 expressing cells after 4-OHT treatment in the MLL-AF9 Ikzf2f/f cre-ER cells. (D-F) All data represent the mean +S.E.M of at least three independent replicates. * p < 0.05, ***p < 0.001, ****p<0.0001 Student’s t test. (G-J) CEBPE depleted cells lacks differentiation induction and IKZF2 targets caused by IKZF2 loss. (G) Differentiation state was measured by flow cytometry using Mac1 and Gr1 as markers in the MLL-AF9 Ikzf2f/f cre-ER cells transduced with lentivirus expressing scramble or Cebpe shRNA and were treated with 4-OHT or left alone. (H) Median Fluorescent Intensity of Mac1 was measured in same experiment in (G). (I) S100a8 (J) S100a9, targets of IKZF2 were measured using qPCR in experiments in (G). (K) Cebpe and (L) Ikzf2 mRNAs were measured to check for depletion in experiment mentioned in (G). (G-L) All data represent the mean +S.E.M of at least three independent replicates. * p < 0.05, **p < 0.01, ****p<0.0001 Student’s t test. (M) Model showing the role of IKZF2 in regulating chromatin accessibility of differentiation and self-renewal program in LSCs. IKZF2 loss leads to increased C/EBPε and chromatin accessibility of regions in differentiation genes containing C/EBP motifs, thereby turning on myeloid genes. HOXA9 expression and chromatin accessibility of HOXA9 motifs are reduced, turning off the self-renewal genes.