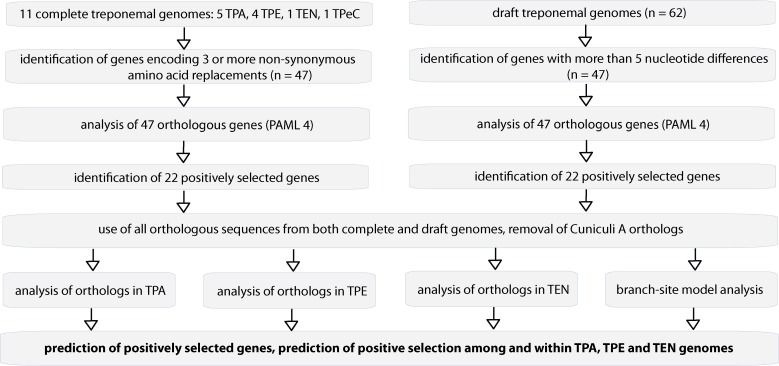
Fig 1. The algorithm used for identification of positively selected genes.
The original search for positively selected genes started with identification of gene orthologs with 3 or more nucleotide differences leading to nonsynonymous amino acid replacements. The original search was performed on a set of 11 complete treponemal genomes listed in Table 1. Subsequently, orthologous gene sequences extracted from published treponemal draft genomes were used and the Cuniculi A orthologs were removed due to frequent sequential diversity and due to lack of pathogenicity of TPeC to humans. Orthologs from draft genomes were used when available and positively selected genes were analyzed within treponemal subspecies using branch-site PAML model analysis.