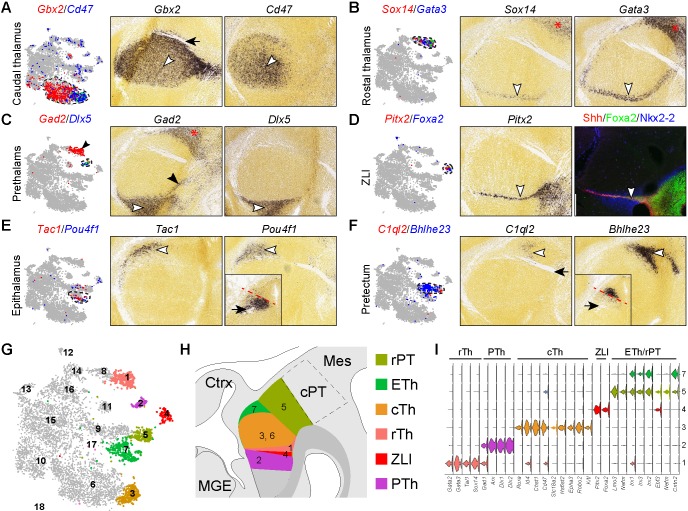
Fig. 2.
Relating postmitotic cell groups to their spatial positions in the diencephalon. (A-F) Expression of cell-specific markers with t-SNE plotting (left) and on sagittal sections of E13.5 mouse diencephalon (right; from the Allen Developing Mouse Brain Atlas). White arrowheads indicate the marker expression domain; black arrow denotes the fasciculus retroflexus; black arrowheads in C show that Gad2 is expressed in the rostral thalamus, as well as the prethalamus; asterisks indicate the caudal pretectum, which exhibits a similar gene expression profile as rostral thalamus. Insets in E and F show the expression of Pou4f1 and Bhlhe23 on coronal sections of E13.5 diencephalon; the red dashed line separates their expression domains. (G) t-SNE projections of diencephalic neurons. (H) Schematic summary of the postmitotic neurons identified by scRNAseq in their endogenous positions in the diencephalon. (I) Violin plot showing expression of genes that are known to mark different diencephalic neurons at E12.5. cPT, caudal pretectum; cTh, caudal thalamus; Ctrx, cortex; ETh, epithalamus; MGE, medial ganglionic eminence; Mes, mesencephalon; PTh, prethalamus; rPT, rostral pretectum; rTh, rostral thalamus; ZLI, zone limitans intrathalamica.